Introduction
miRNAs are a kind of small, endogenous non-coding RNAs that negatively control gene expression at the post-transcriptional level by degrading target mRNAs in plants. To totally make clear the functions of miRNA, it is essential to exactly identify the conserved and species-specific miRNAs and their target genes. Small RNA sequencing and degradome sequencing have been widely used in plants to identify miRNAs and target genes on a genome-wide scale. Many tools have been developed to handle small RNA sequencing and degradome data separately, but no specialized tool exists for a one-stop survey of this kind of interaction.
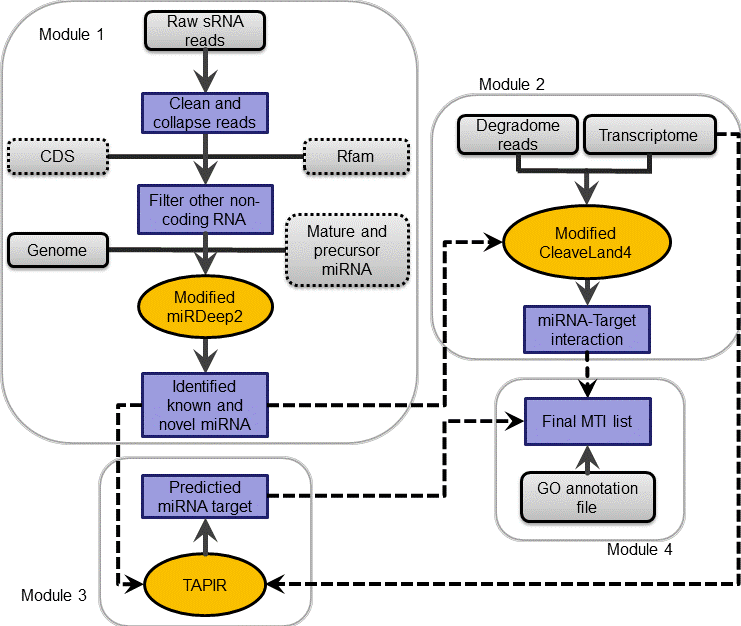
Based on deep sampling of small RNA libraries and degradome fragments of target mRNA by next generation sequencing, MTide enables users to explore expression patterns of known miRNA genes, discover novel ones and identify target mRNA of these miRNAs.
It consists of a modified miRDeep2 with plant-specific parameters, a modified CleaveLand4 with more targets reports and some other useful scripts. To control the running timescale in a low level, we add multiple threads support for miRDeep2, CleaveLand4 and other time-consuming parts.
If you use MTide in your scientific research, please cite us:
Zhao Zhang, Li Jiang, Jingjing Wang, Ming Chen*. MTide: an integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics, doi:10.1093/bioinformatics/btu633.
Implementation
The MTide package was developed in Perl by combining the core algorithm of miRDeep2 and CleaveLand4, cutadapt for adaptor removing, Bowtie for reads mapping and the Vienna RNA package for RNA secondary structure prediction. If users want to prioritize the predicted targets based on GO similarity and do differential expression analysis of miRNA, two R packages csbl.go and DESeq should also be installed. All the scripts can be executed sequentially or by an integrated script MTide.pl in a command line environment. The package has been tested on two Linux platforms, Ubuntu 12.14 and Fedora 8, and should work on similar systems that support Perl and R.
Sequence and Annotation file Downloads
Notice: All the genome don't include chloroplast or mitochondria sequence!
The mature miRNAs and precursor miRNAs are from miRBase release 21. We also provide a script named extract_mirna for extracting mature and precursor miRNA sequences for species which are not listed below!
The GO annotatian files of Ath and Osa are from GO website, and the others are from Gramene. The genome and cdna sequences for Ath and Osa are from Arabidopsis website and MSU, respectively. The sequences for Zma are from Phytozome. The sequences for Gma and Sbi are from Gramene.
Now we only provide files of species which are frequently analysed, and we are trying to collect information of other plant species. If you have any suggestions or questions, please feel free to contact us.
| Species Name | Genome sequence | cDNA sequence | GO annotation | Mature miRNAs | Precursor miRNAs |
|---|---|---|---|---|---|
| Arabidposis thalianna | TAIR10_genome.fa.gz | TAIR10_cdna.fa.gz | ath.go | ath_mature.fa | ath_hairpin.fa |
| Oryza sativa | MSU_v7.0_genome.fa.gz | MSU_v7.0_cdna.fa.gz | osa.go | osa_mature.fa | osa_hairpin.fa |
| Zea mays | Zmays_284_AGPv3.fa.gz | Zmays_284_6a.transcript.fa.gz | zma.go | zma_mature.fa | zma_hairpin.fa |
| Glycine max | Glycine_max.V1.0.23.dna.fa.gz | Glycine_max.V1.0.23.cdna.fa.gz | gma.go | gma_mature.fa | gma_hairpin.fa |
| Sorghum bicolor | Sorghum_bicolor.Sorbi1.23.dna.fa.gz | Sorghum_bicolor.Sorbi1.23.cdna.fa.gz | sbi.go | sbi_mature.fa | sbi_hairpin.fa |
Scripts Downloads
Three downloads are available here:
- MTide.tar.gz includes all the scripts in MTide
- tutorial_data.tar.gz contains the example data
- UserGuide.pdf is the manual file
Contact
For comments or suggestions, please contact us:
Tel/Fax: +86 (0)571-88206122
Zhao Zhang: zhangzhao87@zju.edu.cn
Ming Chen: mchen@zju.edu.cn