Hydra cell atlas Hydra vulgaris Drop-seq 25,052 cells
Study: Stem cell differentiation trajectories in Hydra resolved at single-cell resolution
Stefan Siebert, Jeffrey A. Farrell, Jack F. Cazet, Yashodara Abeykoon, Abby S. Primack, Christine E. Schnitzler, Celina E. Juliano. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science. 2019 Jul 26;365(6451):eaav9314. DOI: 10.1126/science.aav9314.
We generated ~25,000 single-cell transcriptomes from Hydra using Drop-seq, clustered the cells, and annotated cell states. These data identified candidate molecular markers for elusive cell populations such as multipotent interstitial stem cells and germline stem cells. We then constructed differentiation trajectories for cells from each of the three cell lineages found in Hydra using the software URD. This revealed the dynamics of gene expression that occur during cell specification and differentiation in the adult Hydra, including the spatial and temporal expression of transcription factors and gene modules. To identify potential cell state regulators, we used ATAC-seq to reveal regulatory regions of coexpressed genes, identified enriched transcription factor binding motifs within these regulatory regions, and matched these motifs to coexpressed candidate transcriptional regulators. Our trajectory reconstruction also identified similarities between the neurogenesis and gland cell differentiation pathways that suggest a shared or similar progenitor state. We propose a model in which interstitial stem cells give rise to a bipotential progenitor in the ectodermal layer, which crosses the extracellular matrix to supply the endodermal layer with neurons and gland cells. Additionally, trajectory reconstructions of individual cell types (including gland cells and epithelial cells of the ectoderm and endoderm) uncovered gene expression changes that occur as they are translocated along the body column; these results suggest candidate genes and pathways involved in spatial patterning along the oral-aboral axis. Finally, we profiled neurons and neuronal progenitors that were enriched using fluorescence-activated cell sorting and used the data to build a molecular map of the nervous system. We found 12 distinct neuronal subtypes and determined their location using differential gene expression analysis, in situ hybridization, and transgenic approaches. Access to neuronal transcriptional signatures, including the first molecular markers specific to endodermal neurons, creates opportunities for precise manipulations of the nervous system.
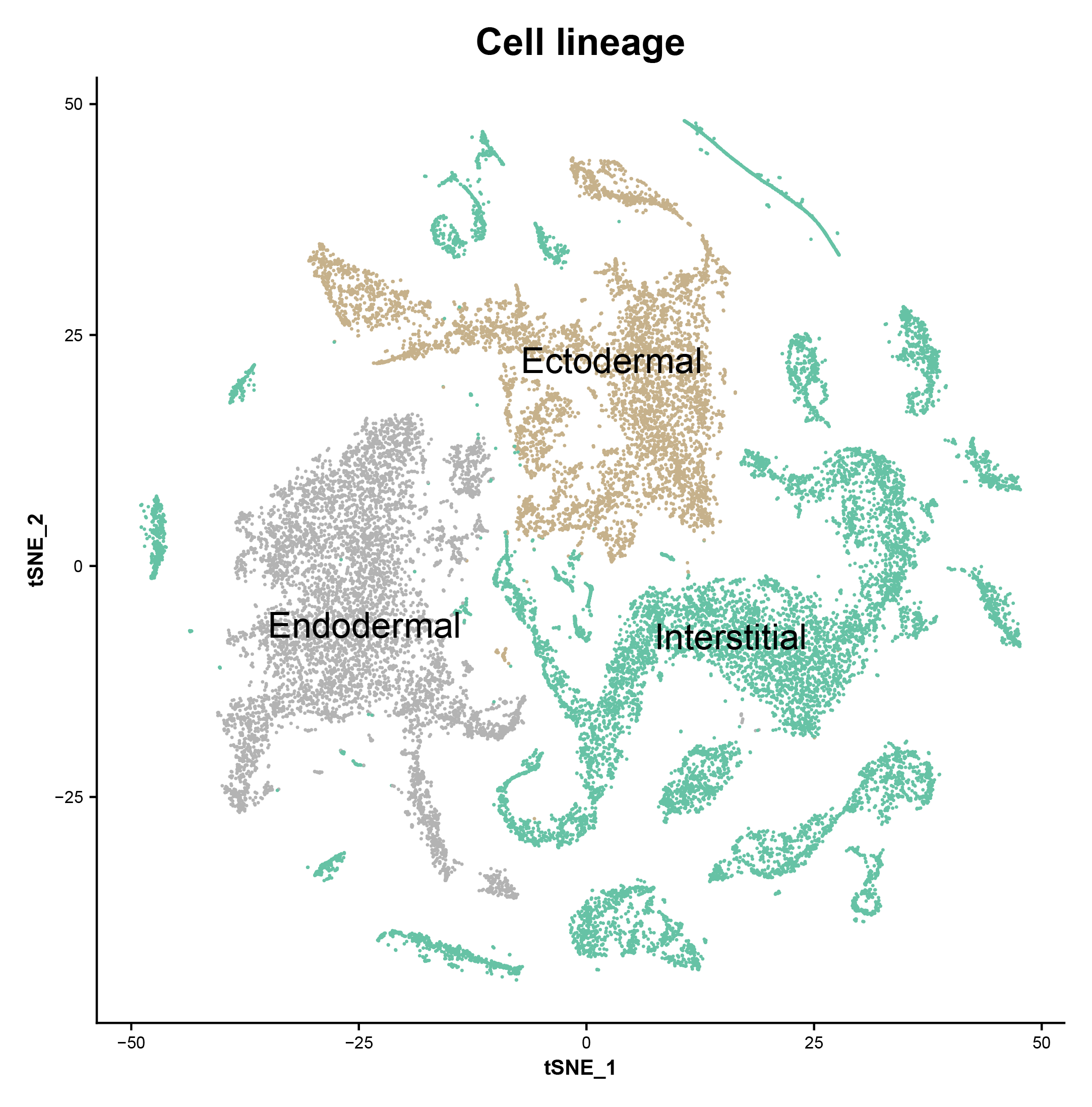
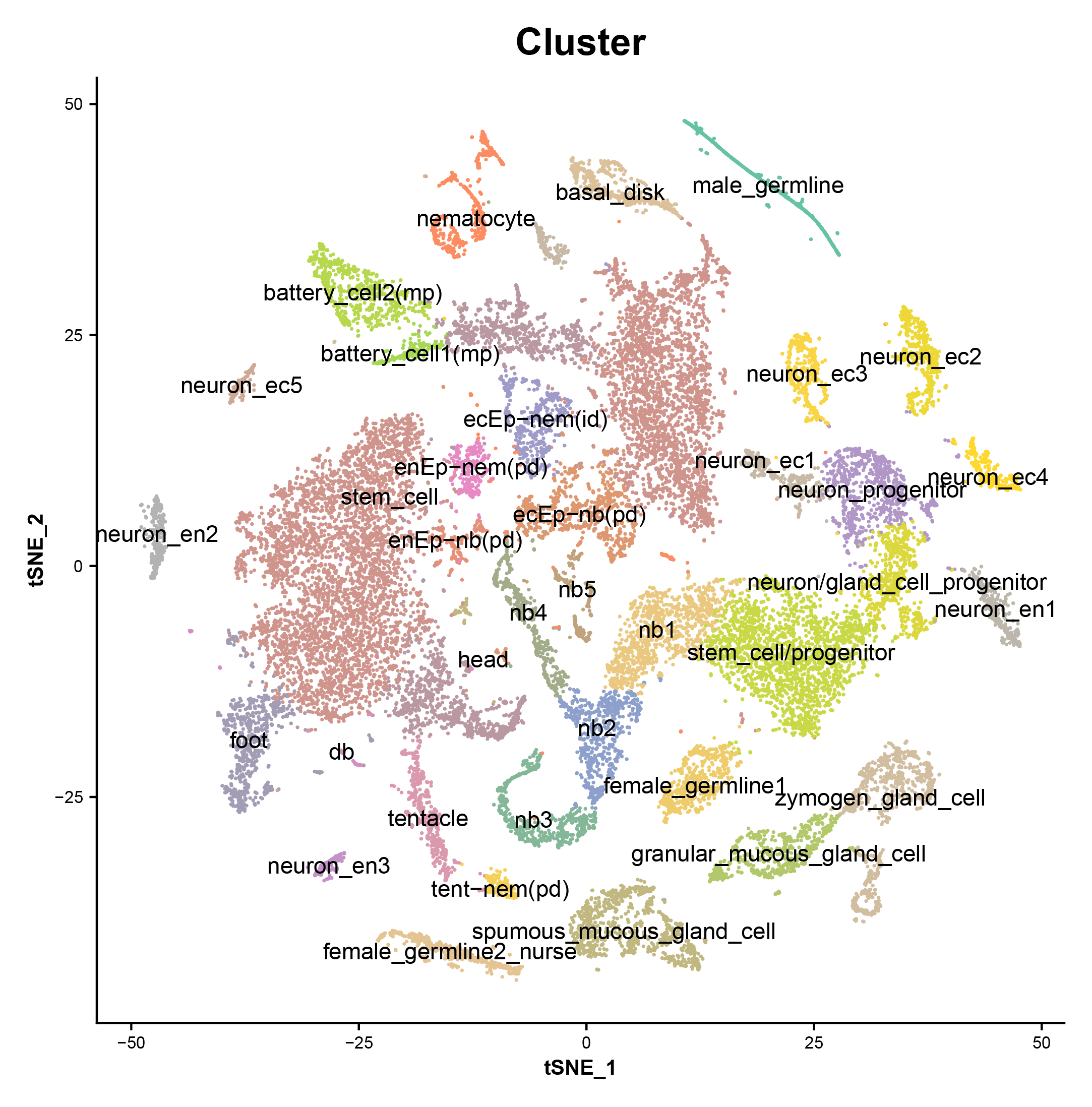

These are marker genes for each cluster.
Study Files
| Filename | Descreption | Download |
|---|---|---|
| Hydra_vulgaris_dge.txt.gz | Expression data of Hydra. | Download |
| Hydra_vulgaris_annotation.txt | Cluster of Hydra. | Download |
Sequence Data
Processed Data
Transcriptome Blast and ATACseq read/peak display
https://research.nhgri.nih.gov/hydra