Reference map of cynomolgus monkeys Macaca fascicularis scRNA-seq 174,233 cells
A reference single-cell regulomic and transcriptomic map of cynomolgus monkeys
Jiao Qu, Fa Yang, Tao Zhu, Yingshuo Wang, Wen Fang, Yan Ding, Xue Zhao, Xianjia Qi, Qiangmin Xie, Ming Chen, Qiang Xu, Yicheng Xie, Yang Sun & Dijun Chen. A reference single-cell regulomic and transcriptomic map of cynomolgus monkeys. Nature Communications. 2022 July 13.
DOI: 10.1038/s41467-022-31770-x.
Non-human primates are attractive laboratory animal models that accurately reflect both developmental and pathological features of humans. Here we present a compendium of cell types across multiple organs in cynomolgus monkeys (Macaca fascicularis) using both single-cell chromatin accessibility and RNA sequencing data. The integrated cell map enables in-depth dissection and comparison of molecular dynamics, cell-type compositions and cellular heterogeneity across multiple tissues and organs. Using single-cell transcriptomic data, we infer pseudotime cell trajectories and cell-cell communications to uncover key molecular signatures underlying their cellular processes. Furthermore, we identify various cell-specific cis-regulatory elements and construct organ-specific gene regulatory networks at the single-cell level. Finally, we perform comparative analyses of single-cell landscapes among mouse, monkey and human. We show that cynomolgus monkey has strikingly higher degree of similarities in terms of immune-associated gene expression patterns and cellular communications to human than mouse. Taken together, our study provides a valuable resource for non-human primate cell biology.

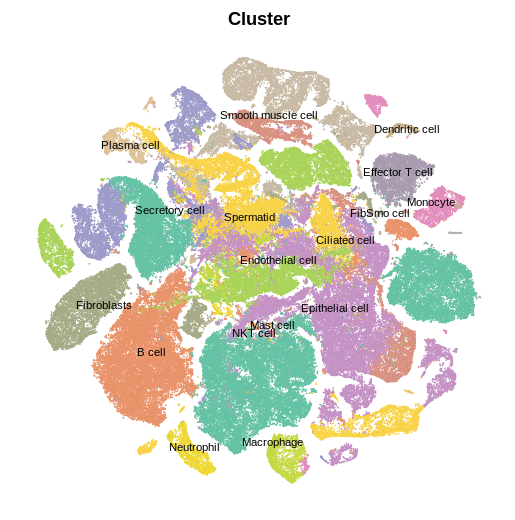

These are marker genes for each cluster.
Study Files
| Filename | Descreption | Download |
|---|---|---|
| Macaca_fascicularis_dge.txt.gz | SCT data of all tissues/organs. | Download |
| Macaca_fascicularis_annotation.txt | Cluster of all tissues/organs. | Download |
Sequence Data
CNP0002427 (scRNA-seq) and CNP0002441 (scATAC-seq)
GSE196792 (scRNA-seq) and GSE196791 (scATAC-seq)