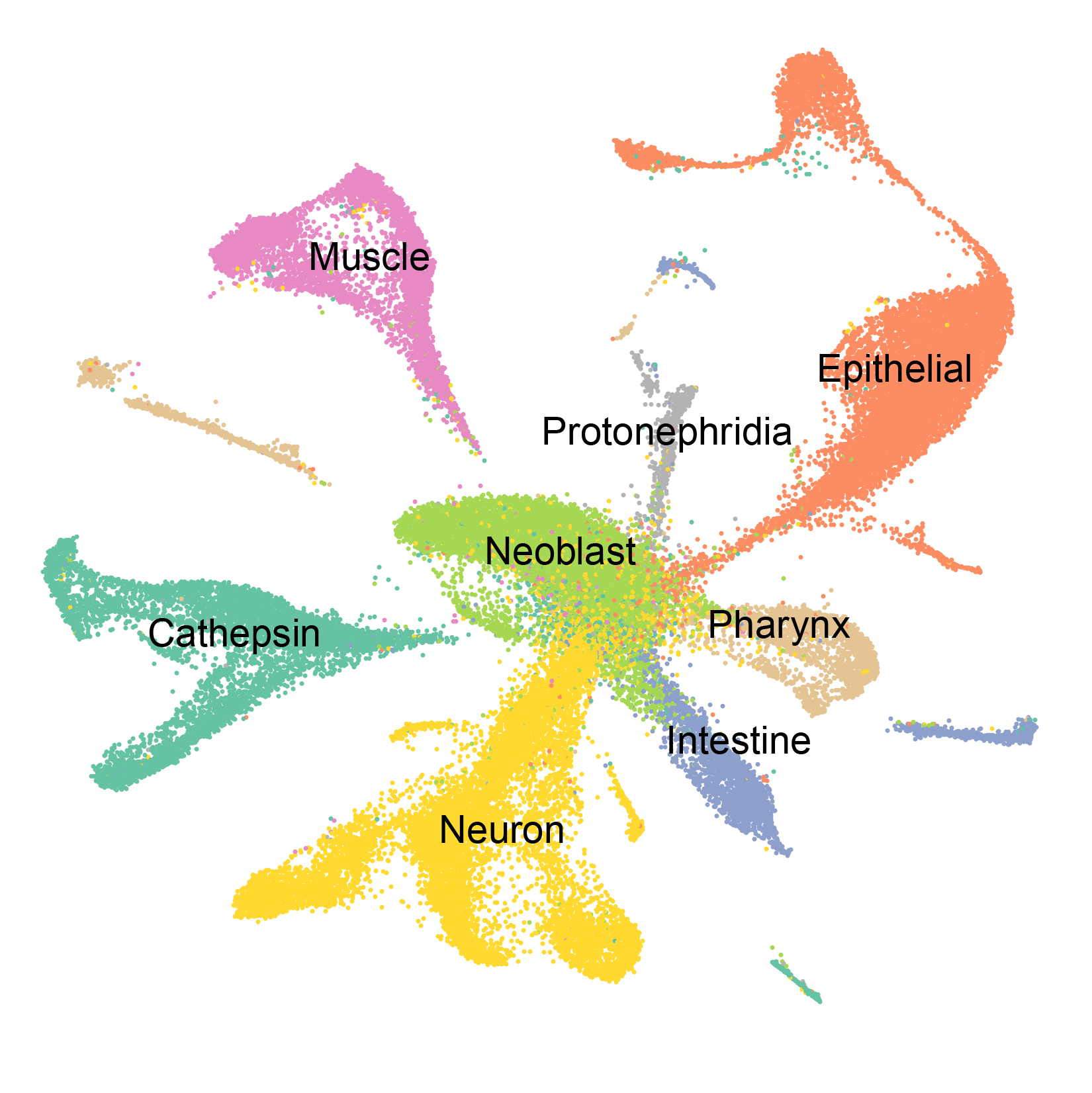
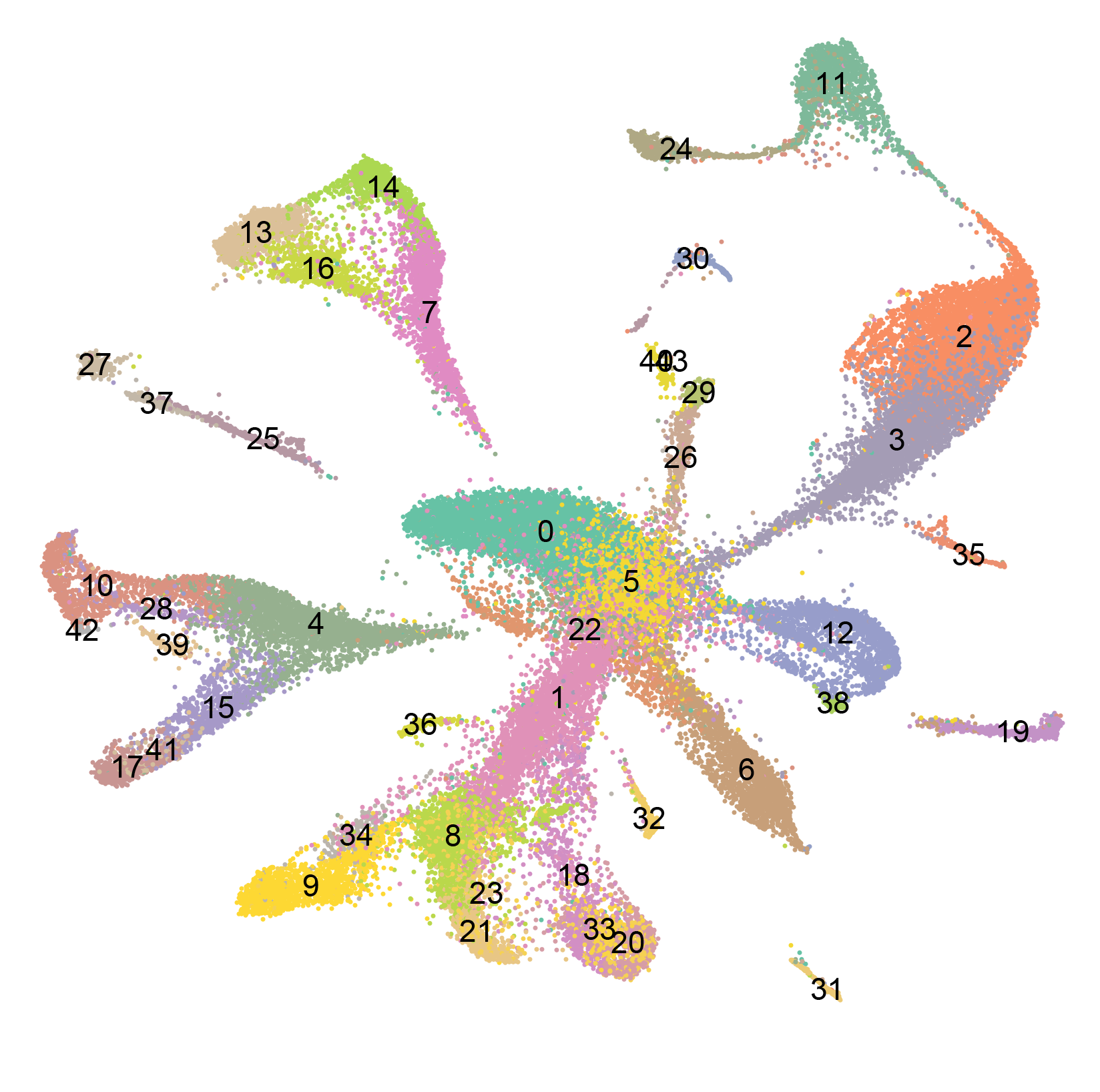
Adult planarian cell atlas Schmidtea mediterranea Drop-seq 66,783 cells
Study: Cell type transcriptome atlas for the planarian Schmidtea mediterranea
Christopher T. Fincher, Omri Wurtzel, Thom de Hoog, Kellie M. Kravarik, Peter W. Reddien. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science. 2018 May 25;360(6391):eaaq1736.
DOI: 10.1126/science.aaq1736.
We used the SCS method Drop-seq to determine the transcriptomes for 66,783 individual cells from adult planarians. We locally saturated cell type coverage by iteratively sequencing distinct body regions and assessing the frequency of known rare cell types in the data. Clustering the cells by shared gene expression grouped cells into broad tissue classes. Subclustering of each broad tissue type in isolation enabled separation of cells into the cell populations constituting each tissue. These analyses enabled the identification of a previously unidentified tissue group and the classification of poorly characterized tissues into their constituent cell types, including numerous previously unknown cell types. Transcriptomes were identified for many rare cell types, including those that exist as rarely as ~10 cells in an animal that has 105 to 106 cells, which suggests that near-to-complete cellular saturation was reached. In addition, transcriptomes for known and novel lineage precursors, from pluripotent stem cell to differentiated cell types, were generated. Precursor transcriptomes identified transcription factors required for maintenance of associated differentiated cells during homeostatic cell turnover. Finally, the data were used to identify genes regionally expressed in muscle, which is the site of planarian patterning gene expression.