About HPInet
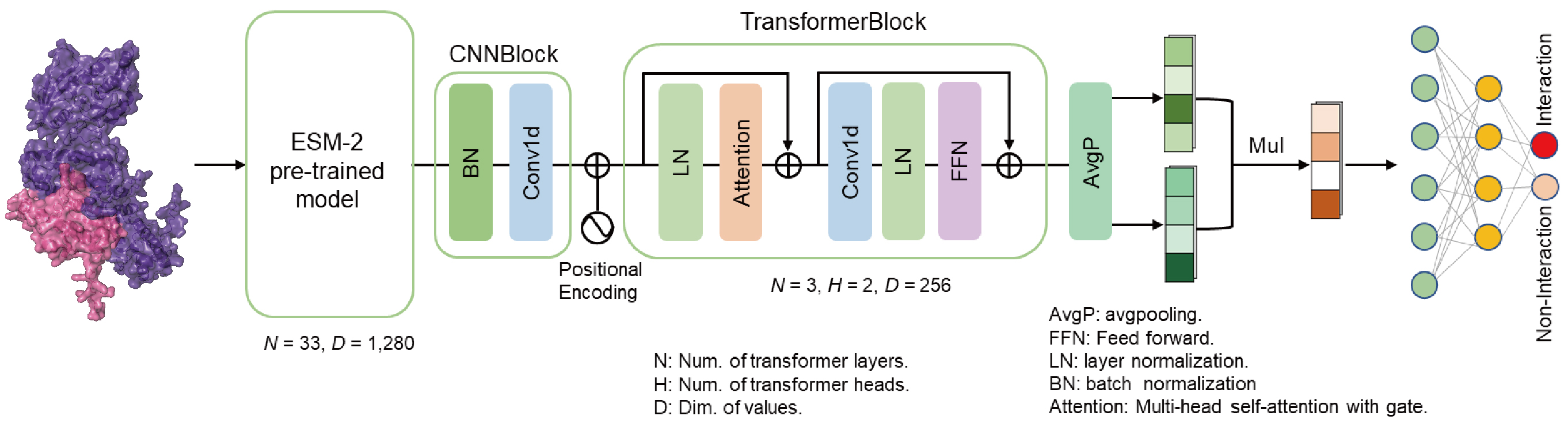
Architecture of the HPInet model.
The HPInet classifier is a deep learning model based on a siamese network architecture, designed for predicting protein-protein interactions (PPIs). The core of the model is a transformer module that integrates convolutional feature extraction and global response normalization (GRN), multi-head self-attention mechanisms to perform protein feature representation learning and classify interaction relationships. If you would like to learn more about our model, feel free to refer to our article.
Network
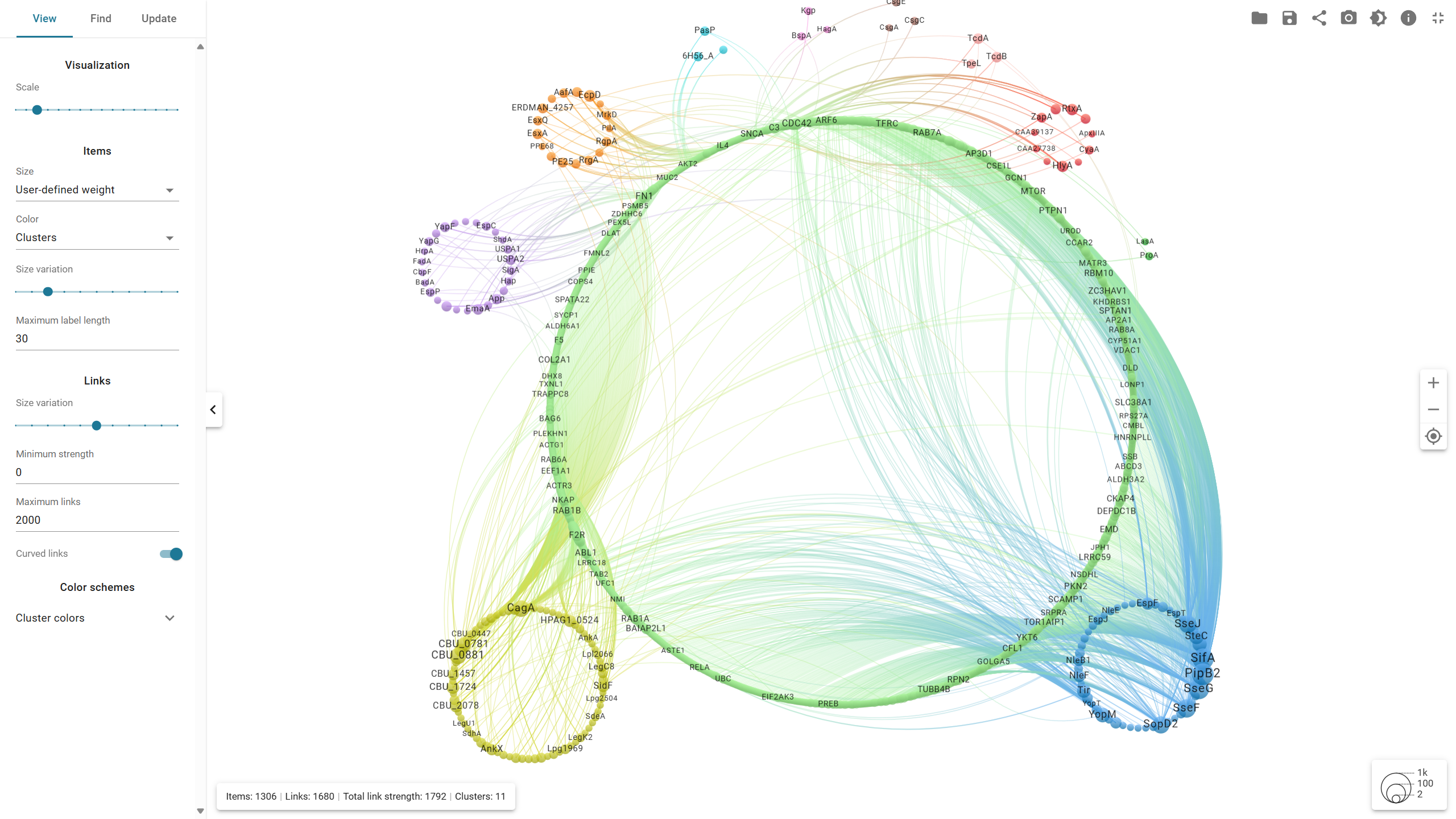
Interaction network of human proteins and bacterial effector proteins.
This figure is dynamically displayed on the homepage, allowing users to customize the graphical parameters using VOSviewer control panel for further research. The interactions cover all secretion systems (T1SE-T10SE), with the majority originating from T3SE and T4SE, accounting for 1,202 and 379 interactions, respectively. The edges represent protein-protein interactions (PPIs), while the size of each node reflects the number of interactions associated with each protein.
How to Use the Prediction Tool
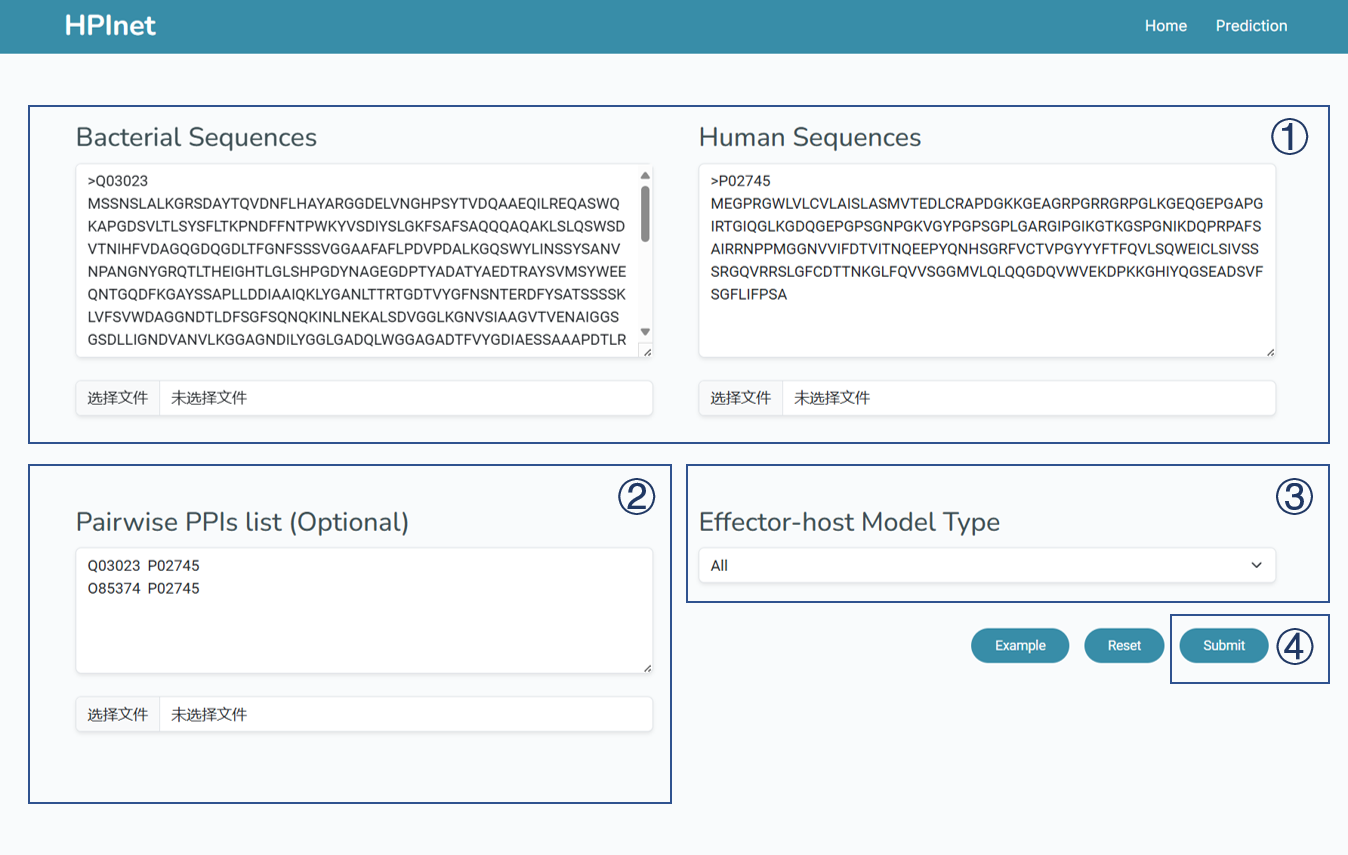
How to use the HPInet model web server.
To use the online HPInet model web server, follow these steps:
- Upload or input your protein sequences in FASTA format.
- Upload or input a PPI list (optional).
- Select the model type you need. There are now four secretion protein models (T3SE, T4SE, T5SE, T7SE) and one general model available for selection.
- Click the "Submit" button to run the model.
The model will then analyze the uploaded sequences and generate predictions for each pair. The results will be displayed in the specified output format. Once loading is complete, the page will show a table of predicted interacting protein pairs along with a diagram of the interactions. Users can click on any row in the table to view detailed information about that interaction.
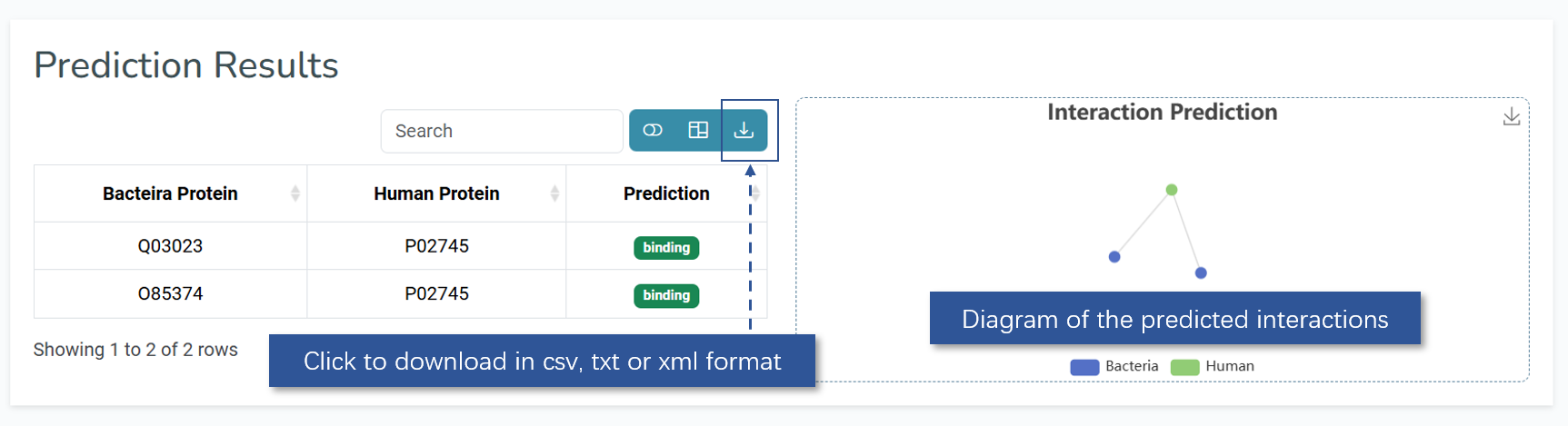
Simplified result of the HPInet model displayed in prediction page.
Result
In the detailed result page, each pair of interacting proteins will display the following scores for both the bacterial protein and the human protein:
- Importance Scores.
- Transmembrane helices
- Signal peptides
- Subcellular Location