Introduction
The ncPlantDB is a comprehensive database designed to facilitate research and discovery in the field of plant non-coding RNAs (ncRNAs) and non-coding peptides (ncPEPs). Our database provides extensive information on various ncRNA-related entities, supporting researchers worldwide in their quest for understanding plant genomics and molecular biology.
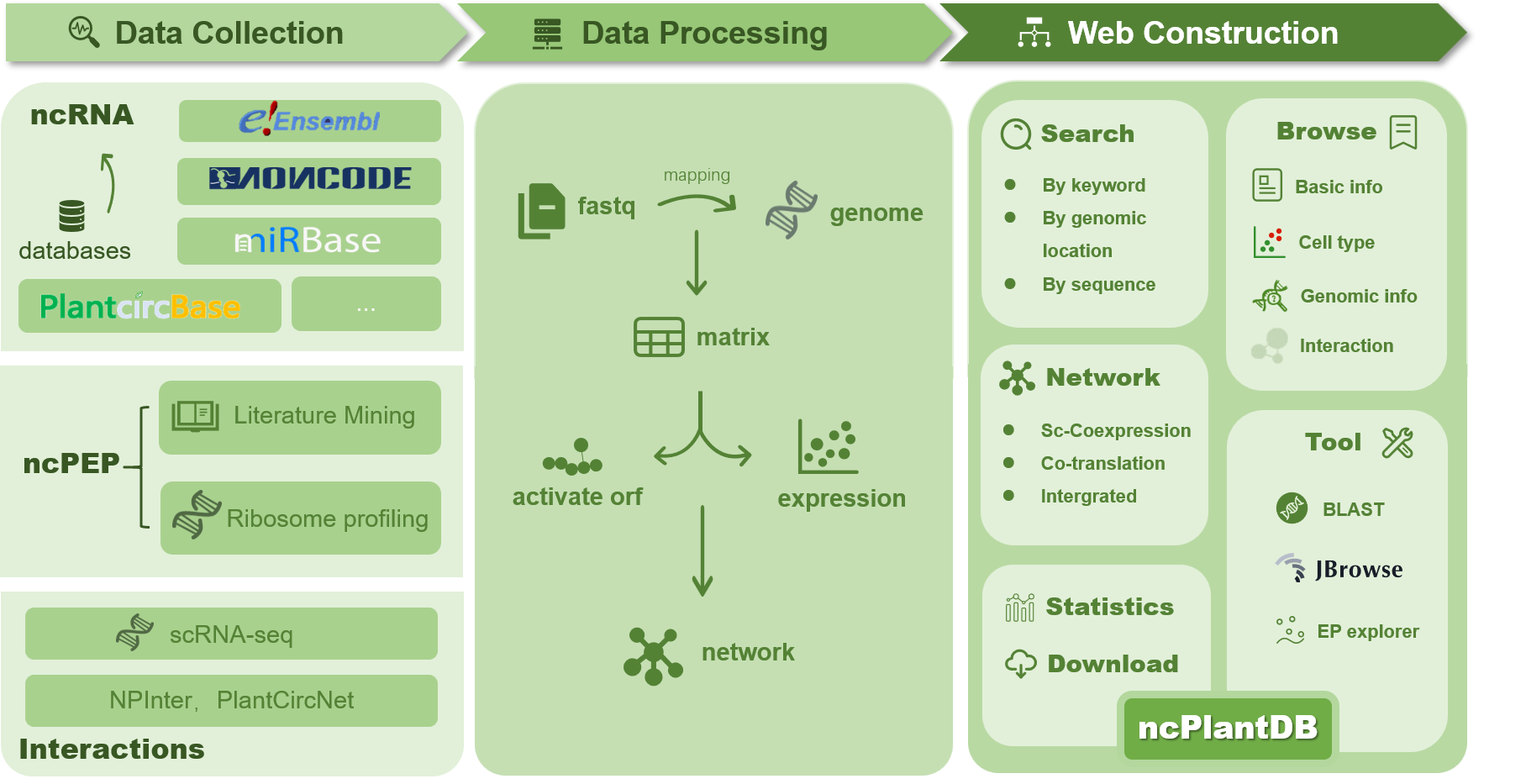
Key Features
- Translational Potential Data: ncPlantDB includes data on ncRNAs with potential coding sequences and experimentally validated functional peptides, providing a unique resource for researchers interested in the translational aspects of ncRNAs.
- Celltype-Specific Interaction Networks: Our database offers detailed ncRNA interaction networks from a cell-specific perspective, helping researchers understand the complex interactions at the cellular level.
- Co-Translational Interaction Networks: ncPlantDB also includes information on co-translational interaction networks, providing insights into the interactions that occur during the translation process.
- User-Friendly Interface: The database features a robust and intuitive interface that allows users to easily search and analyze data by keywords, genomic locations, and sequences.
- Comprehensive Search Results: The search results provide detailed information about the queried entities, including identifiers, names, types, genomic coordinates, and other relevant data, supporting in-depth analysis and research.
- Interactive Tools: ncPlantDB includes interactive tools for viewing and analyzing search results, such as the Expression Pattern Explorer, which allows users to analyze gene expression patterns across different tissues and conditions.
Search
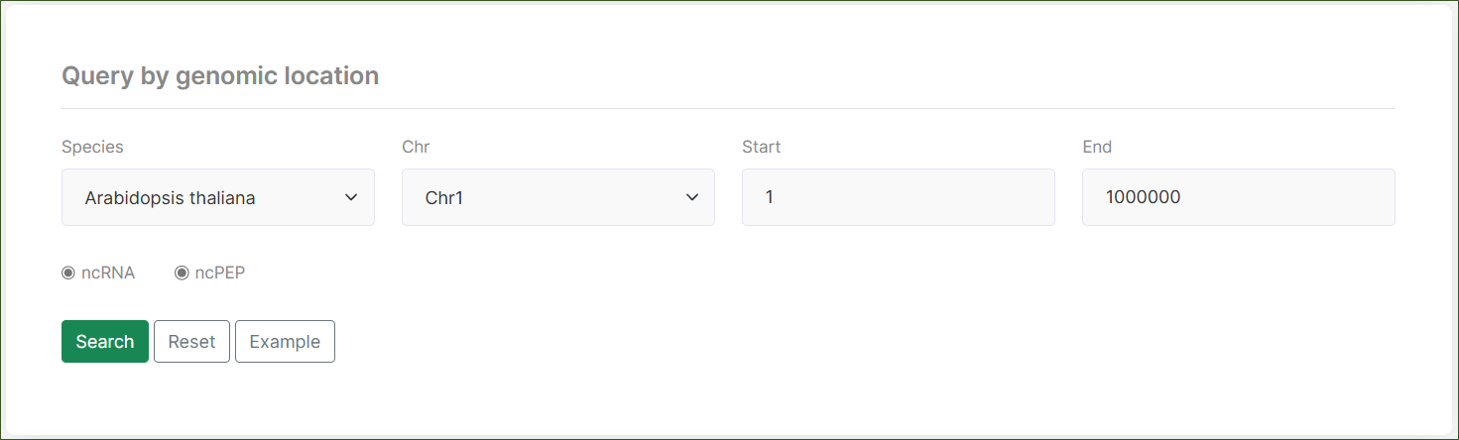
The ncPlantDB provides a comprehensive search interface designed to help users quickly find specific ncRNA-related data. Our search functionality is divided into three main sections:
Query by Keyword:
This section allows users to search for ncRNAs, ncPEPs, or interactions using specific keywords or identifiers. Users can select the species of interest and enter the locus ID or name to retrieve relevant data. The search results provide detailed information about the queried entities.
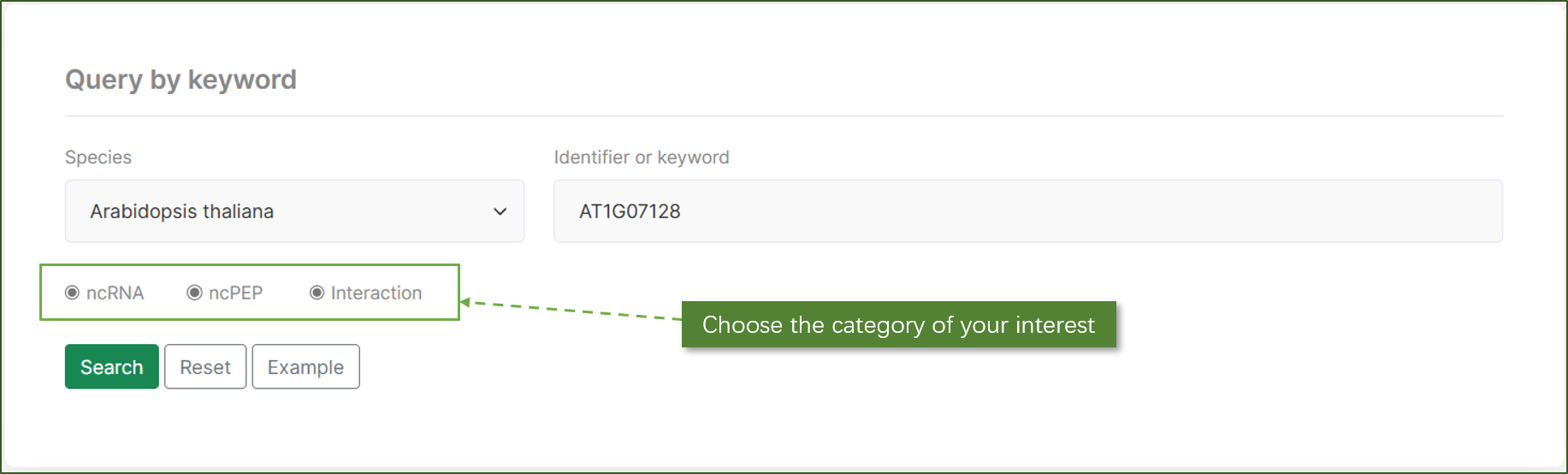
Query by Genomic Location:
Users can search for ncRNAs or ncPEPs based on their genomic coordinates. By specifying the species, chromosome, and start and end positions, users can locate specific genomic regions of interest. This functionality is particularly useful for researchers focusing on specific genomic loci.
Results
The results for both keyword and location searches are presented in a structured format.Users can click on the "detail" icon to navigate to the detailed page for more in-depth information.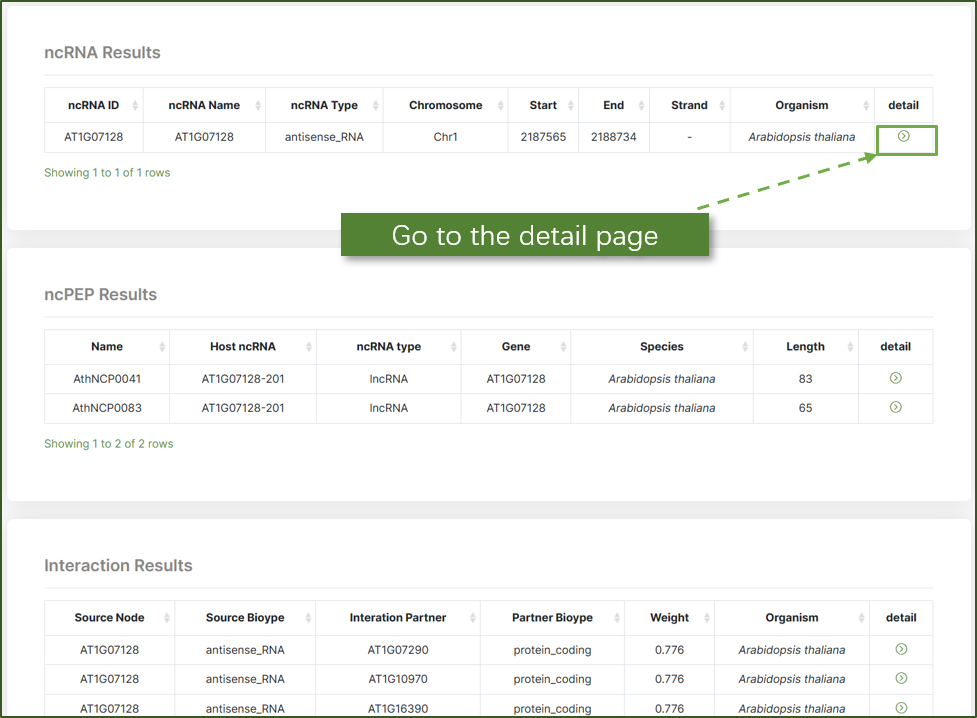
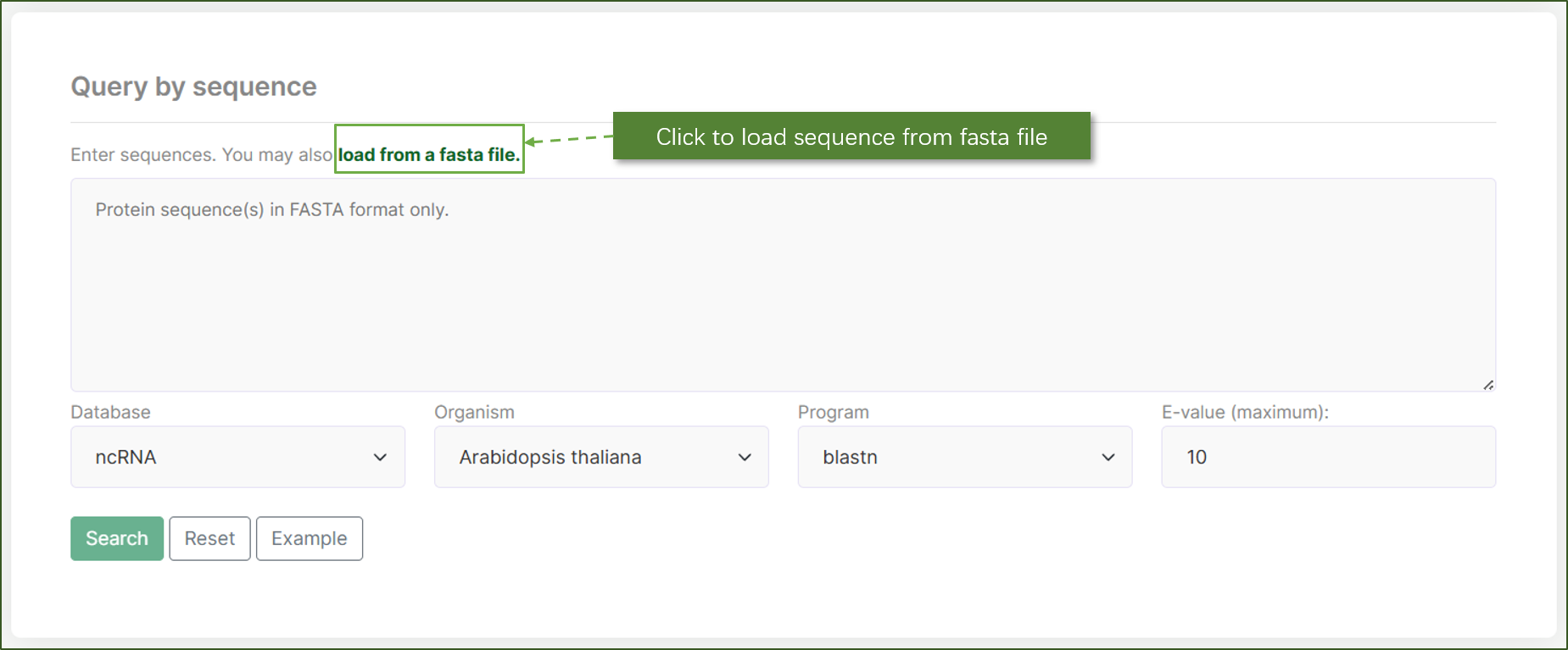
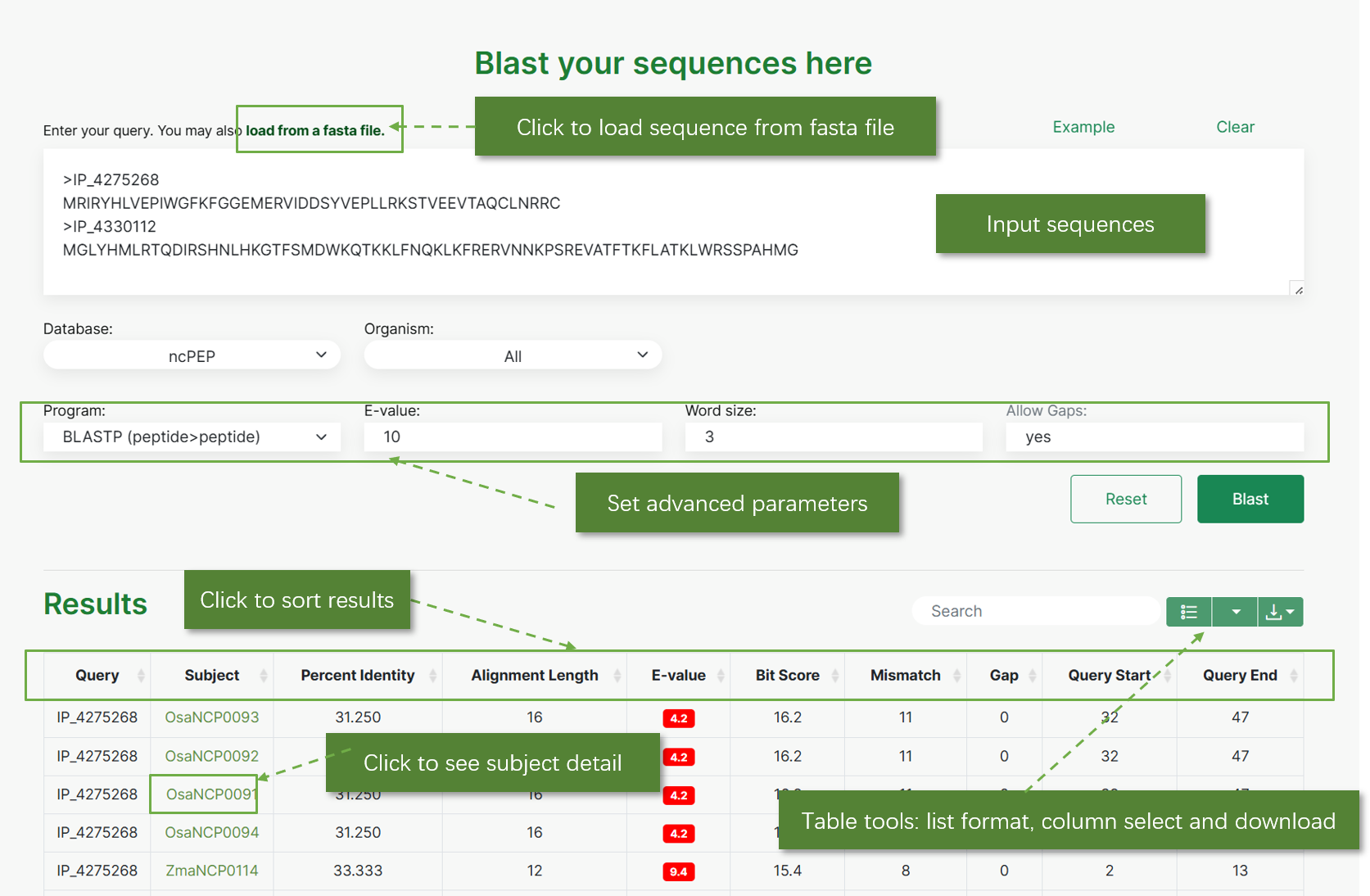
Query by sequence:
This section enables users to search for ncRNAs by inputting sequences in FASTA format using NCBI BLAST. Users can load sequences from a FASTA file or enter them directly into the search box. The interface allows selection of the database, organism, and search program (e.g., BLASTN), along with setting the maximum E-value for sequence alignment results.
Results
The BLAST results display a comprehensive list of alignments. Users can click on the subject identifier to view detailed information about the alignment. The table tools allow users to change the list format, select columns, and download the results for further analysis. NCBI out formats are also available.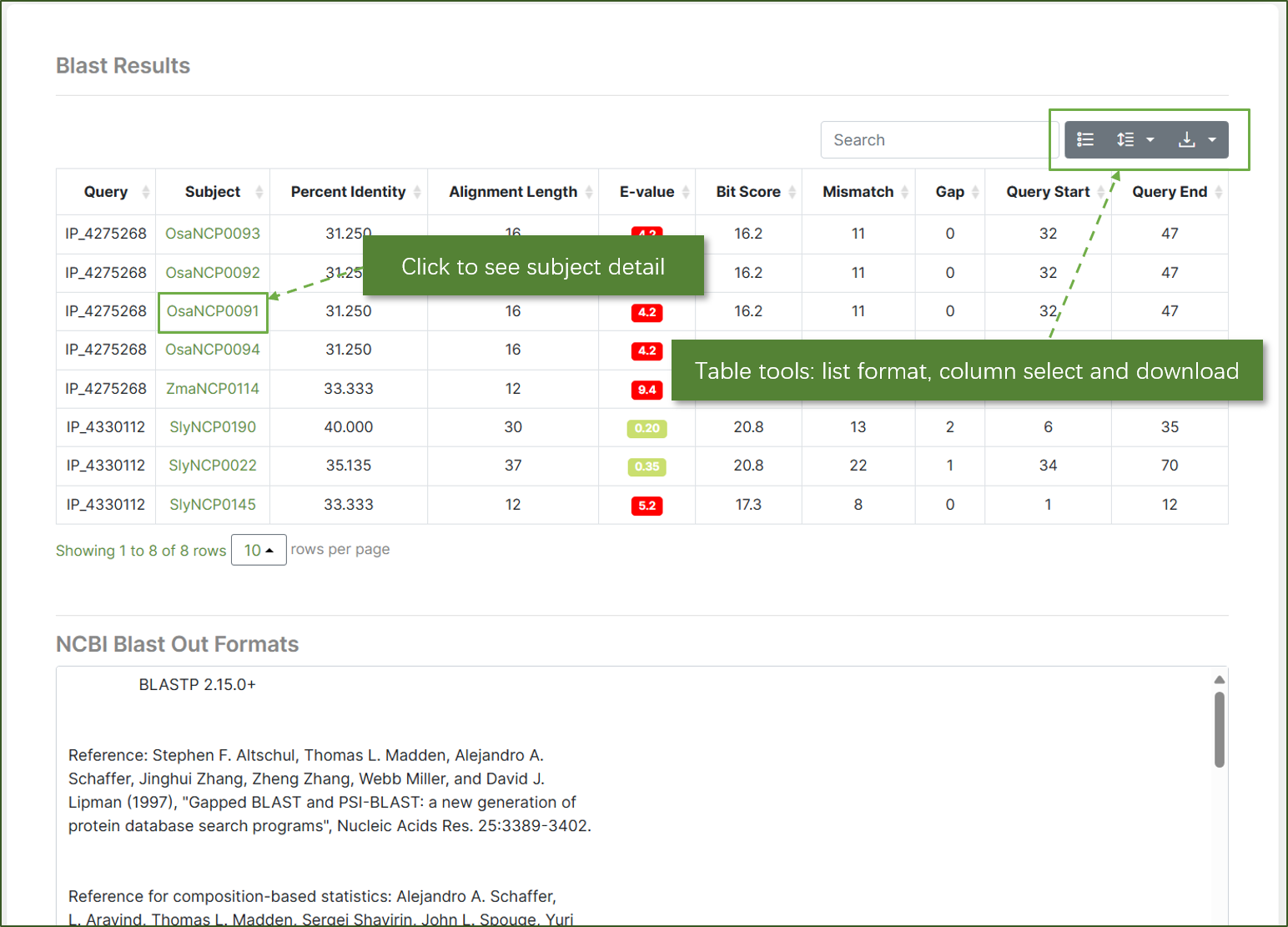
Our search interface is designed to be intuitive and user-friendly, ensuring that researchers can efficiently access the specific data they need. By providing multiple search options, ncPlantDB caters to a wide range of research requirements, facilitating in-depth analysis and discovery in the field of plant ncRNAs.
Browse
At ncPlantDB, we provide a user-friendly interface for browsing our database. You can explore the ncRNA database by filtering and searching for specific ncRNAs. Our browsing section is categorized into three main parts:
-
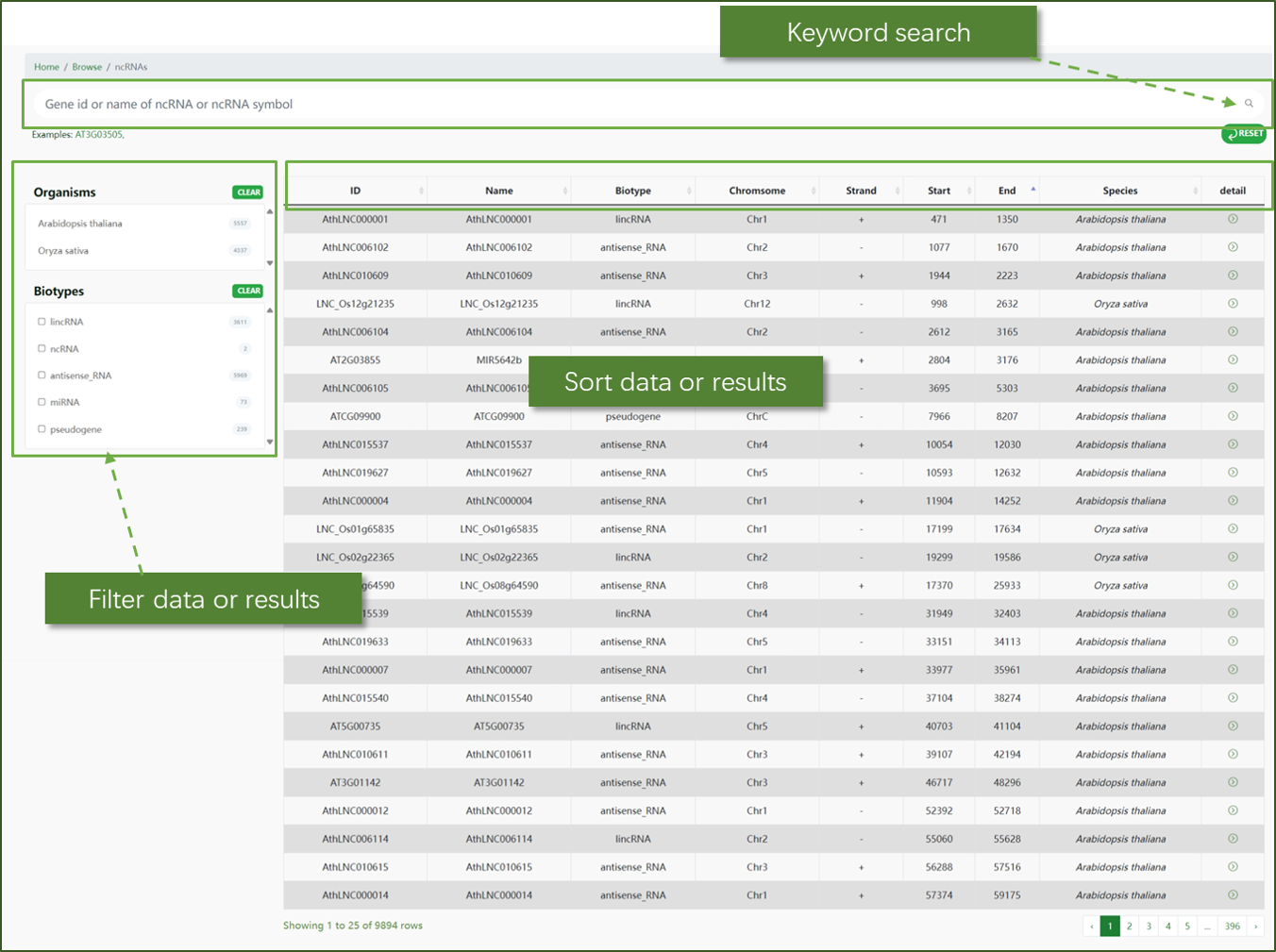
ncRNA:
This section allows users to browse through various non-coding RNAs. You can filter and search for specific ncRNAs based on different criteria such as species, biotype, and chromosomal location. The table view provides detailed information including ID, name, biotype, chromosome, strand, start and end positions, and species.
-
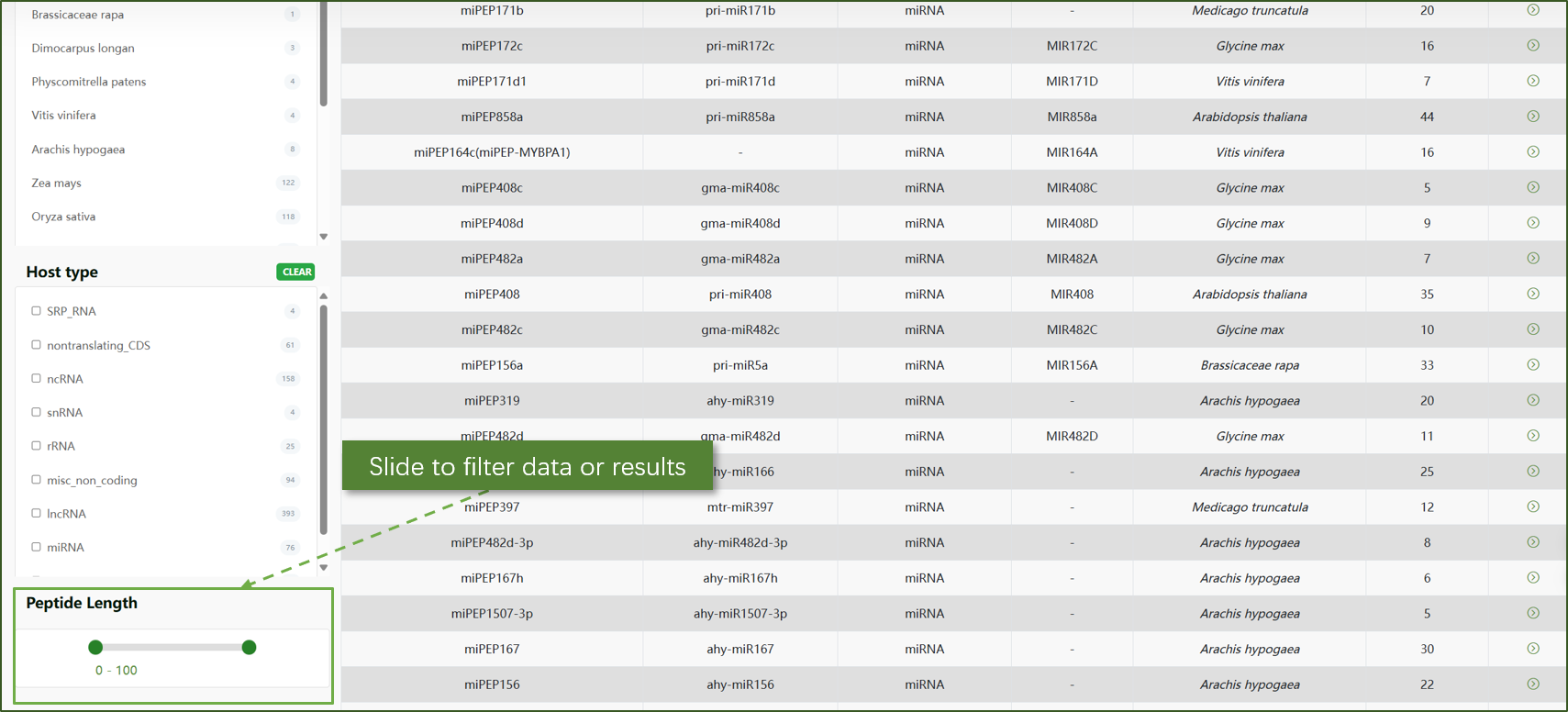
ncPEP:
Here, users can explore non-coding peptides that are derived from ncRNAs. This section provides detailed information about the sequences, annotations, and potential functions of these peptides. You can filter and search for specific ncPEPs based on criteria such as species, host type, and peptide length. The table view includes columns for name, host ncRNA, ncRNA type, gene, species, and peptide length.
Interaction:
This part of the database focuses on the interactions between ncRNAs and other molecules. Users can browse through interaction data to understand the regulatory networks and pathways involving ncRNAs. The table view includes detailed information such as source node, source biotype, interaction partner, partner biotype, weight, organism, cell type, and data source.
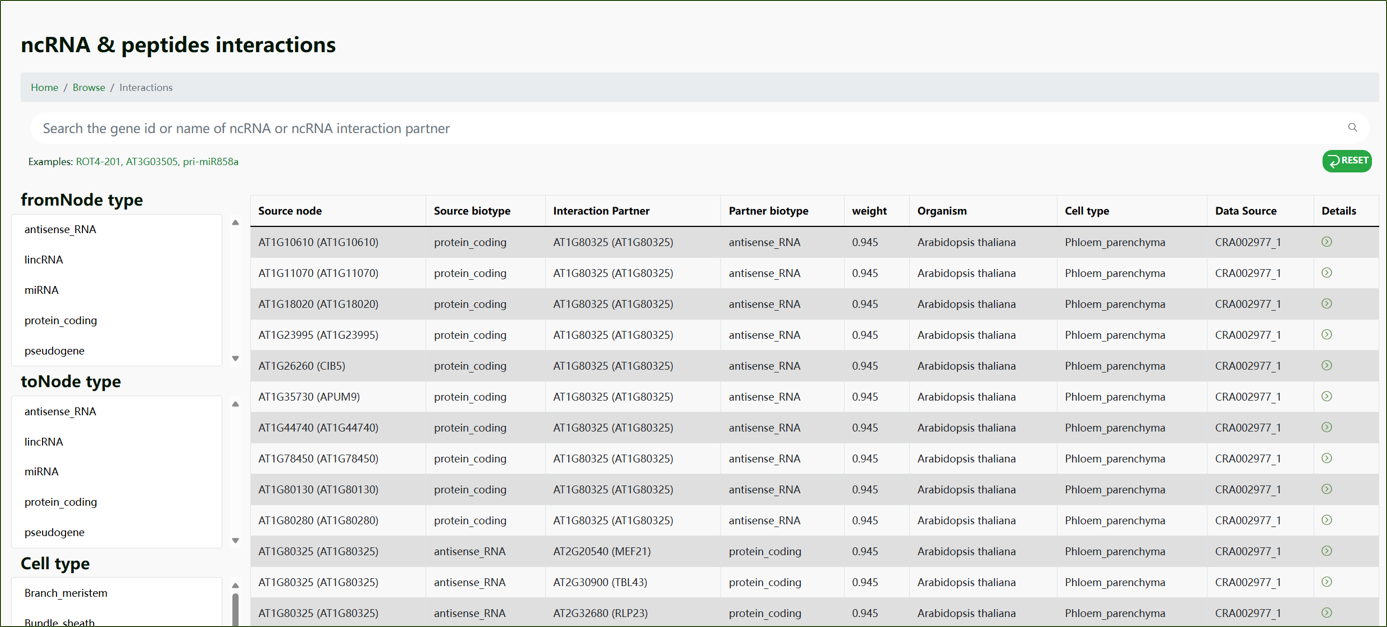
By providing these categorized browsing options, we aim to make it easier for researchers to navigate through the vast amount of data available in ncPlantDB. Our user-friendly interface ensures that you can quickly find and access the specific information you need for your research.
Network
At ncPlantDB, we aim to provide comprehensive information on plant ncRNAs, including their interactions with other ncRNAs and molecules. Our network section focuses on these interactions, offering valuable insights into the regulatory roles of ncRNAs.
Currently, we have developed two types of Weighted Gene Co-expression Network Analysis (WGCNA) networks:
-
Single-cell Level ncRNA WGCNA Network:
This network is constructed based on single-cell expression data, allowing users to explore the co-expression patterns of ncRNAs at a highly detailed cellular resolution. It is invaluable for understanding the cell-specific regulatory roles of ncRNAs.
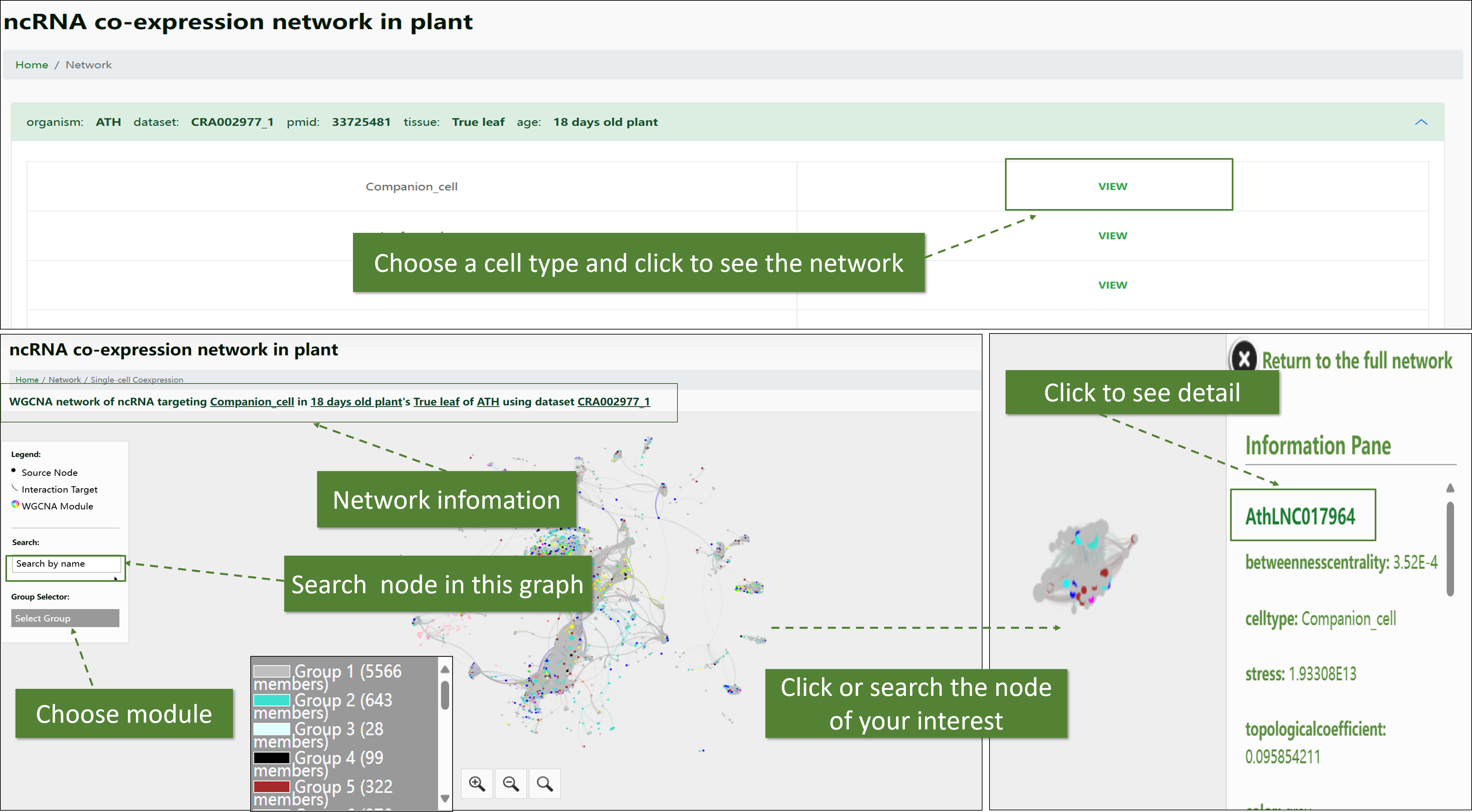
-
Translational Level WGCNA Network (Co-translational Network):
This network is derived from Ribo-seq expression matrices, focusing on the interactions at the translational level. It highlights the co-expression relationships of ncRNAs during the translation process, providing insights into their roles in protein synthesis and post-transcriptional regulation. Most functions are the same as Single-cell Level ncRNA WGCNA Network.
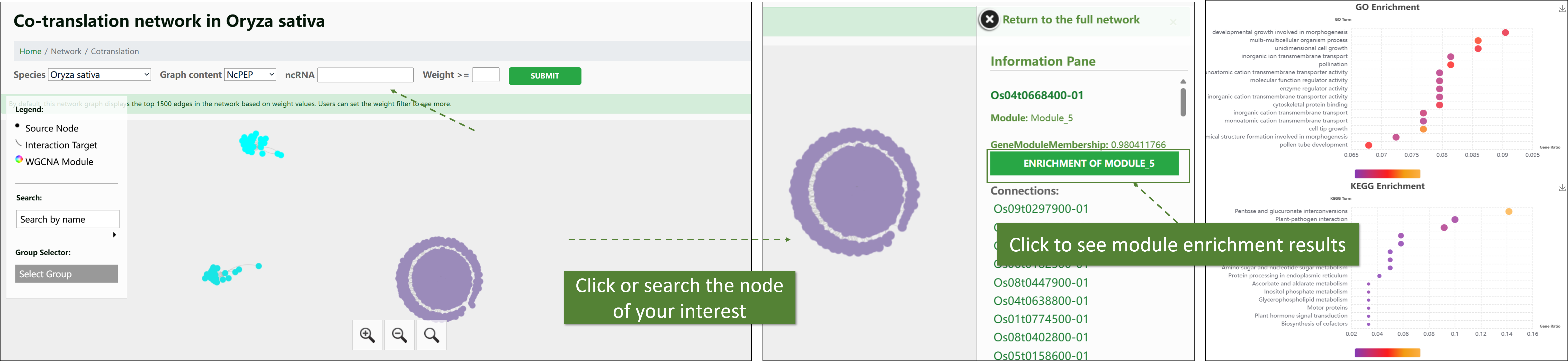
-
Integrated Hub Network:
This network is based on the Single-cell Level ncRNA WGCNA Network. The hub genes were extracted based on their contribution to the network. Additionally, this network integrates information about ncRNAs that encode ncPEPs (non-coding peptides). This provides a comprehensive view of the central players in the ncRNA regulatory landscape, offering insights into their multifunctional roles at both the cellular and translational levels.
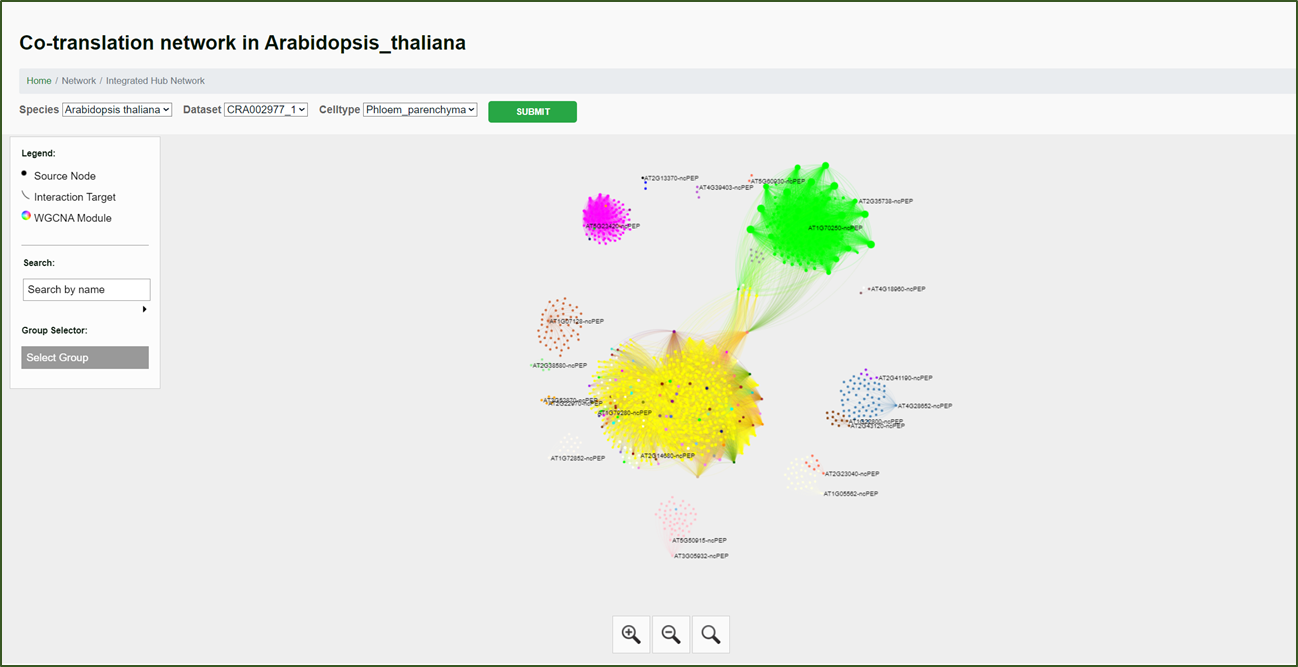
Moreover, we have integrated these networks to form a comprehensive hub gene network. This feature allows researchers to identify key regulatory ncRNAs (hub genes) that play central roles in various biological processes and pathways.
Our network section is one of the unique features of our website, offering a powerful tool for researchers to delve into the intricate web of ncRNA interactions. By providing these detailed and specialized networks, ncPlantDB supports the exploration of ncRNA functions and their regulatory mechanisms in plants.
Tools
We provide tools to help you explore and analyze plant ncRNAs and ncPEPs. Currently, we offer two primary tools:
-
BLAST:
The Basic Local Alignment Search Tool (BLAST) allows users to compare their sequences against our comprehensive database to identify regions of similarity. This is essential for understanding the functional and evolutionary relationships of ncRNAs.
-
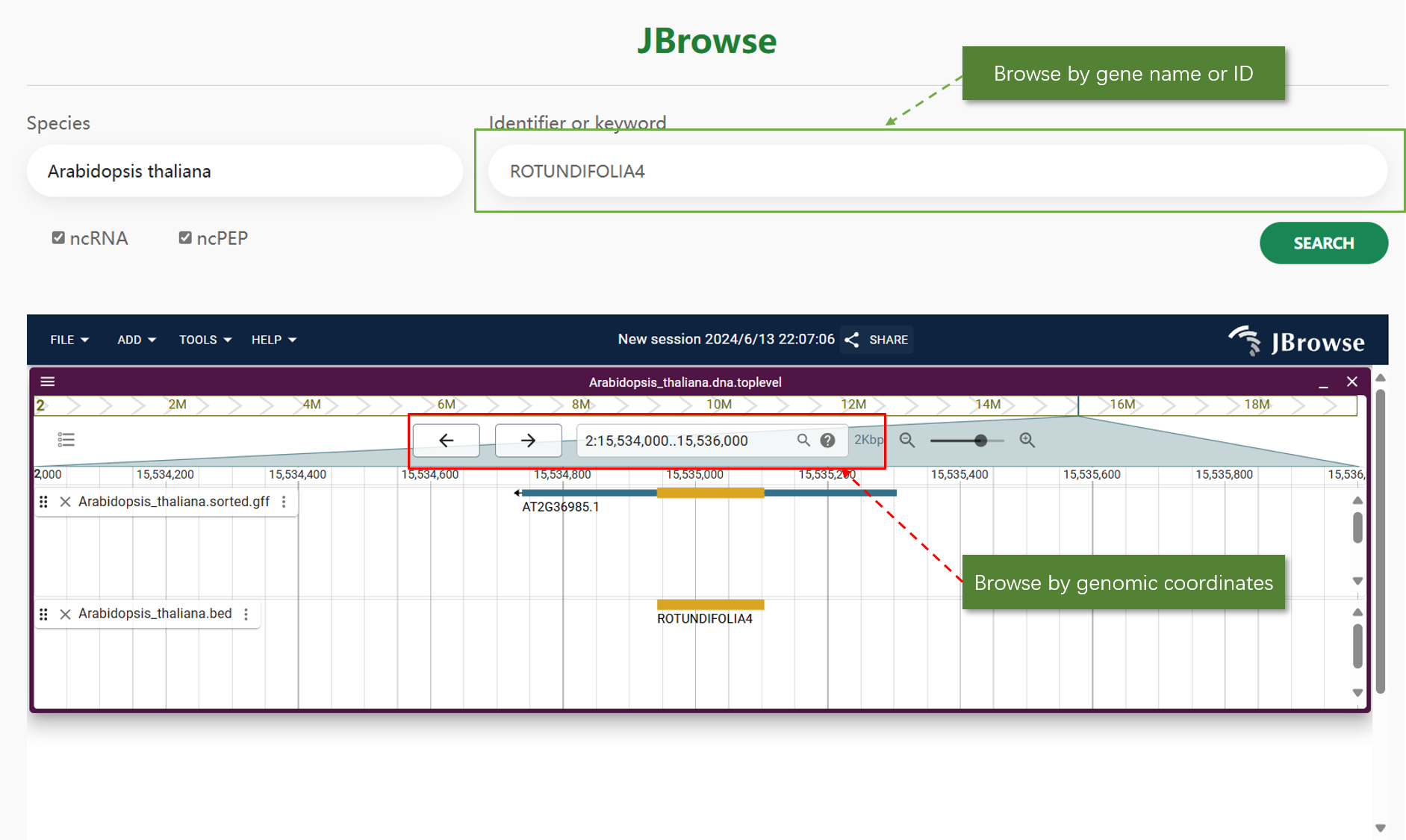
JBrowse:
JBrowse is a genome browser that provides an interactive interface for visualizing genomic data. Users can easily navigate through different genomic regions, view annotations, and examine the structural features of ncRNAs and ncPEPs. The web tool allows users to identify ncRNA regions by keyword search.
-
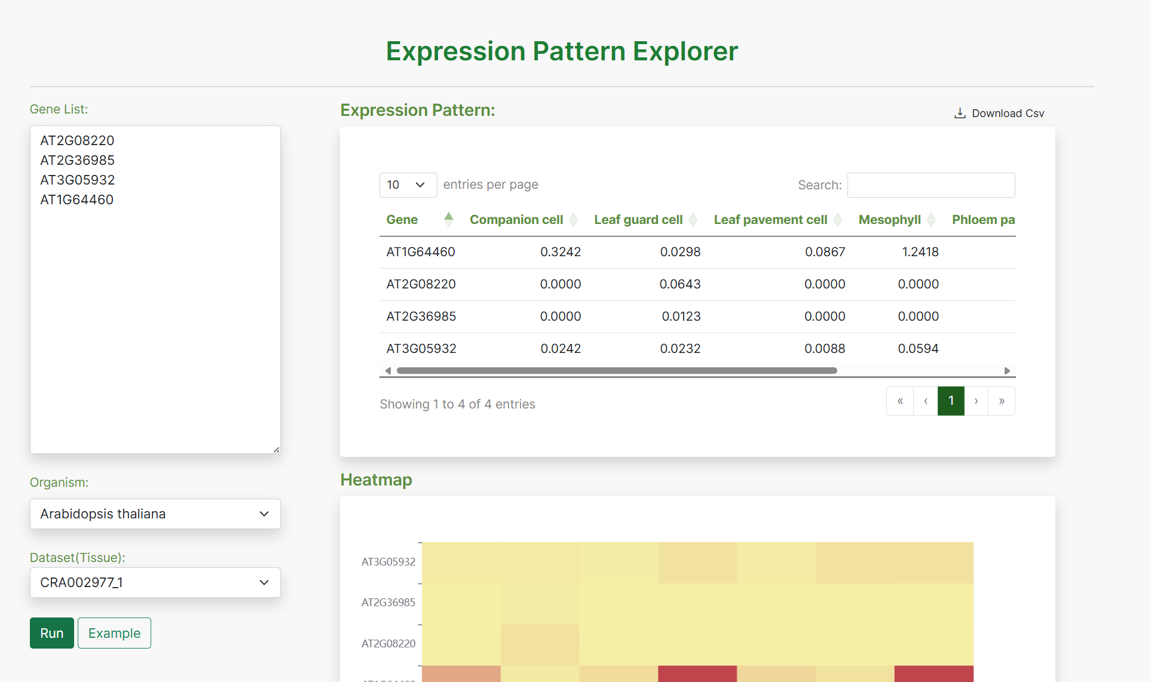
Expression Pattern Explorer:
The Expression Pattern Explorer (EPE) is a tool that allows users to visualize the expression patterns of ncRNAs across different tissues and celltypes. Users can input a list of gene IDs, select the organism and dataset (tissue), and run the analysis to retrieve expression data. The results include an expression pattern table and a heatmap.
Submit
Welcome to submit plant ncRNAs or ncPEPs to ncPlantDB!
Your submission will be appreciated! Please email your ncRNAs or ncPEPs to 3190103446@zju.edu.cn with the following information:
- Organism (required)
- Genomic location (required)
- Strand (required)
- Sequence (required)
- Name
- Other information
Contact
We value your insights and are eager to address any questions or comments you may have. You can find our contact details here. We look forward to hearing from you and engaging in meaningful conversations.
Ming Chen's lab, College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, China
Ø Telephone: +86 (0)571-88206612
Ø Fax: +86 (0)571-88206612
Ø E-mail: Ming Chen (mchen@zju.edu.cn); Liya Liu (3190103446@zju.edu.cn)
Download
We have provided resources in a variety of formats to help you explore the data, including FASTA, GFF, and CSV.
Users can download these valuable resources based on their species of interest. The resources are primarily divided into three categories:
-
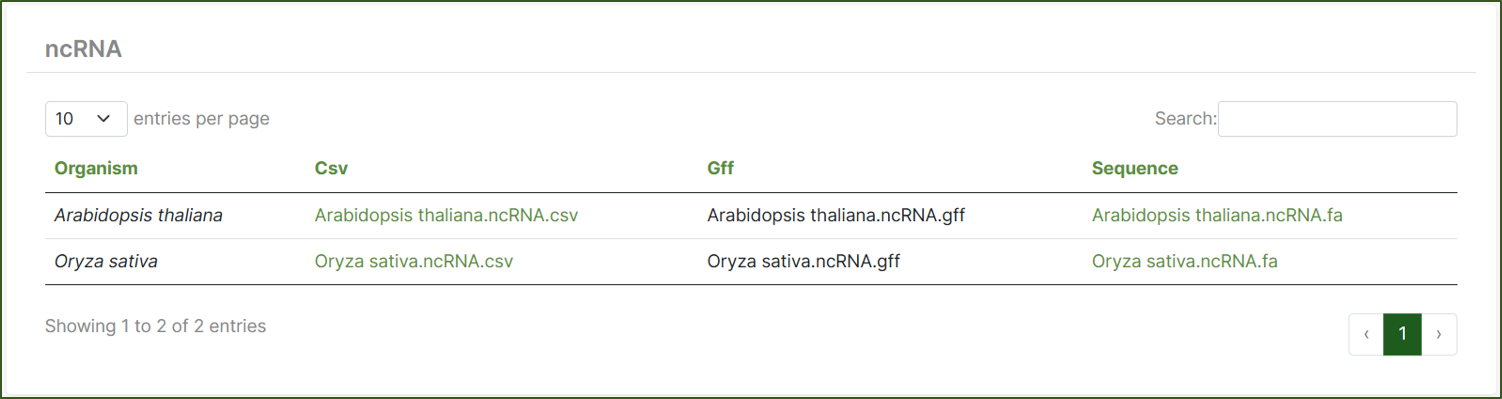
ncRNA:
This category includes nucleotide sequences and annotations of non-coding RNAs. Available in FASTA, GFF, and CSV formats, these datasets are essential for sequence analysis, functional studies, and genomic research.
-
ncPEP:
This section provides data on non-coding peptides, which are derived from ncRNAs. The datasets are available in various formats, including FASTA for sequence data and CSV for tabular information, facilitating the study of their roles and functions.
-
Interaction Data:
This category includes data on the interactions between ncRNAs, as well as between ncRNAs and other molecules. There are two sections in this category: one for Single-cell Level ncRNA WGCNA Network and another for Translational Level WGCNA Network (Co-translational Network). The datasets are available in CSV format and zipped by species.
By offering these different formats and categories, we ensure that researchers have the tools they need to effectively explore and analyze plant ncRNA data. Whether you are interested in sequence alignment, genomic annotation, or interaction studies, our download section provides the necessary resources tailored to your specific research requirements.