
Welcome to PceRBase!
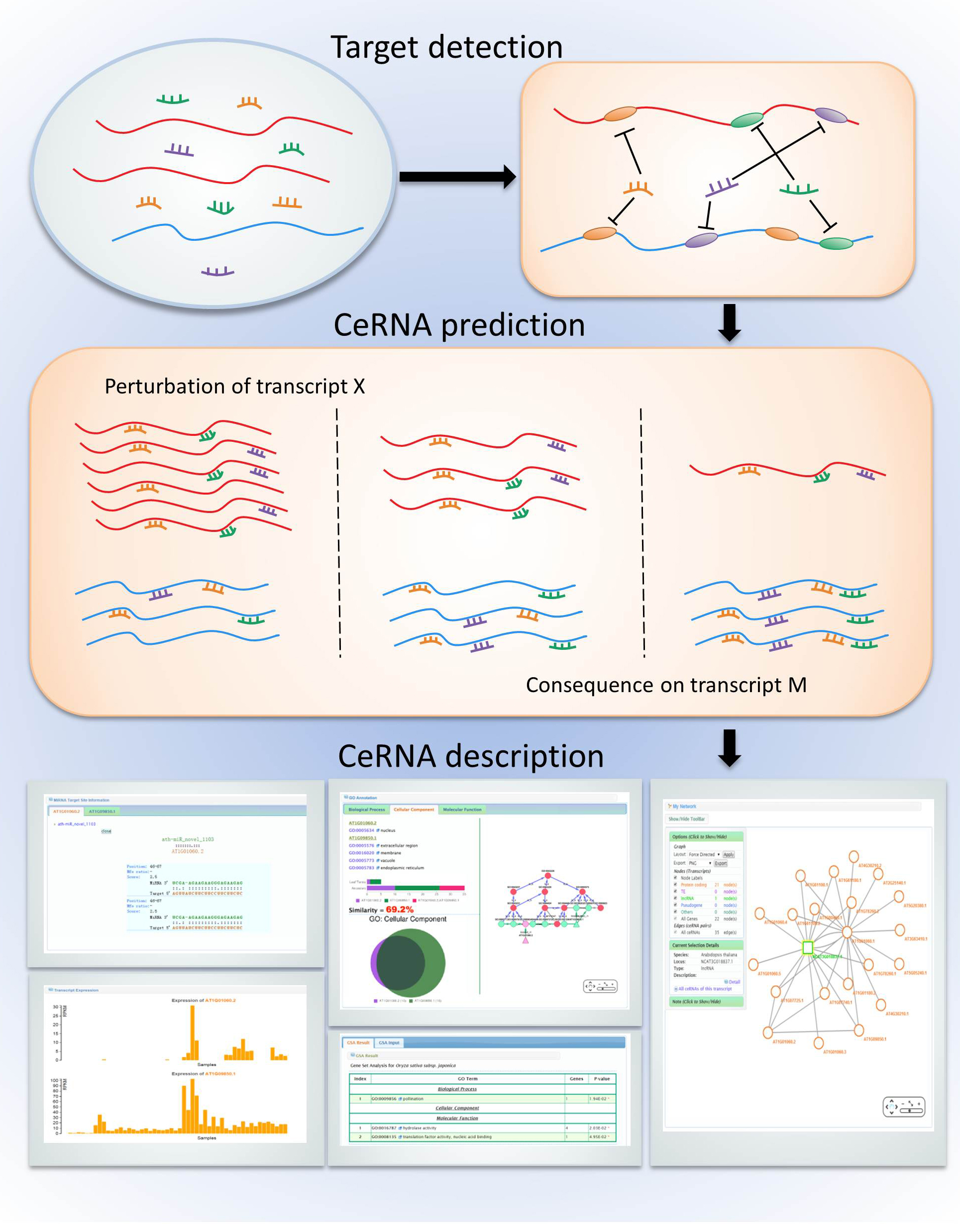
Competing endogenous RNA (ceRNA) act as decoys for microRNA (miRNA) binding, thereby regulating the abundance of other RNA transcripts which share the same microRNA response elements (MREs).
Plant ceRNA database (PceRBase) uses mRNA, long non-coding RNA and miRNA sequence information to predict potential
The likelihood of the interaction between these pairs of candidate ceRNAs is further tested by considering co-expression of predicted ceRNAs. Predicted pairing structure between miRNAs and their target mRNA transcripts, expression levels of ceRNA pairs and associated GO annotations are also stored in the database.
PceRBase currently predicts 696,756 potential ceRNA target-target, and 167,608 ceRNA target-mimic pairs from 28 plant species. Particularly, 239,998 ceRNA target-target and 75,360 ceRNA target-mimic pairs were found in Arabidopsis thaliana.
Users can submit their own sequence information and expression data to PceRBase to predict significant ceRNA interactions and their biological significance.
Update News:
Add B.napus, H.annuus to the database.
2016-06-08
Public Z.mays lncRNA datasets were integrated into PceRBase.
2016-05-27
PceRBase website construction finished.
2016-03-05
The “Gene Set Analysis” module was integrated into
PceRBase.
The GO annotation was provided by PceRBase.
2015-10-20
The “Predict” web-tool was integrated into PceRBase.
2015-09-08
Add 24 new plant species to database.
2015-07-19
The PceRBase website was initially constructed.
2015-6-21
The ceRNA prediction of Arabidopsis and rice finished.
All rights reserved by Ming Chen's Lab, Zhejiang University.