Cell type-specific screening results

Epithelial

|
|
C0_Epithelial cell (malignant)
C1_Enterocyte
C2_AT2 cell
C3_Epithelial cell
C4_Acinar cell
C5_Enterocyte
C6_Goblet cell
C7_Proliferating epithelial cell (malignant)
C8_Exocrine cell
other 14 cell types
|
Endothelial

|
|
C0_Tip EC (cancer-associated)
C1_Venous EC (cancer-associated)
C2_Capillary EC
C3_Sinusoidal EC_FTL high
C4_Sinusoidal EC_STAB high (cancer-associated)
C5_Arterial EC (cancer-associated)
C6_Immune-related EC (cancer-associated)
C7_Lymphatic EC
C8_EC_SFTP high
C9_EC_SPINK1 high
|
Myeloid

|
|
C0_Monocyte_CXCL2 high
C1_Macrophage_INHBA high
C2_Macrophage_STAB1 high
C3_cDC_CD1C high
C4_Monocyte_HSP high
C5_Mast cell
C6_Neutrophil_CD55 high
C7_Macrophage_THBS1 high
C8_Macrophage_SFTPC high
other 9 cell types
|
Stromal

|
|
C0_Fibroblast_MGST1 high
C1_Myofibroblast (cancer-associated)
C2_Vascular smooth muscle cell (cancer-associated)
C3_Fibroblast_CXCL14 high (cancer-associated)
C4_Fibroblast_FBLN1 high (cancer-associated)
C5_Pericyte (cancer-associated)
C6_Smooth muscle cell
C7_Fibroblast_CCL19 high (cancer-associated)
C8_Fibroblast_CCL5 high (cancer-associated)
other 2 cell types
|
Lymphoid

|
|
C0_Tm cell
C1_CD8 T cell
C2_Plasma cell_IGHG high
C3_Plasma cell_IGHA high
C4_NK cell
C5_B cell
C6_Plasma cell_MT high
C7_Treg cell
C8_Proliferating T cell
other 3 cell types
|
Other

|
|
C1_Unknown
C7_Unknown
C8_Schwann cell
C9_Other
C9_Proliferating cell
C10_Schwann cell
C12_Other
C14_Proliferating cell
C15_Proliferating cell
C16_Schwann cell
other 2 cell types
|
Apply Shennong framework to the pan-cancer landscape and explored the response of the tumor cells to pharmacologic compounds.
Introduction of the Shennong framework
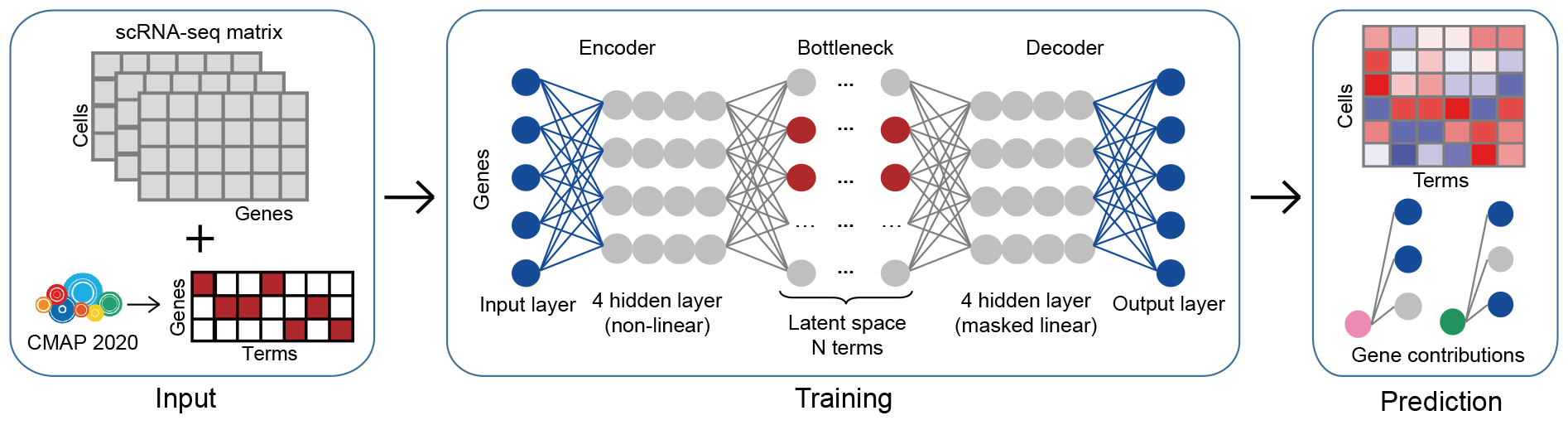
Shennong (https://github.com/PeijingZhang/Shennong/) is a deep learning framework for in silico screening of anticancer drugs for targeting each of the landscape cell clusters. Utilizing Shennong, we could predict individual cell responses to pharmacologic compounds, evaluate drug candidates’ tissue damaging effects, and investigate their corresponding action mechanisms. Prioritized compounds in Shennong’s prediction results include FDA-approved drugs currently undergoing clinical trials for new indications, as well as drug candidates reporting anti-tumor activity. Furthermore, the tissue damaging effect prediction aligns with documented injuries and terminated discovery events. This robust and explainable framework has the potential to accelerate the drug discovery process and enhance the accuracy and efficiency of drug screening.
Processed count matrices and cell annotations are available at figshare: https://doi.org/10.6084/m9.figshare.25497445.
Citation: Peijing Zhang†, Xueyi Wang†, Xufeng Cen†, Qi Zhang†, Yuting Fu, Yuqing Mei, Xinru Wang, Renying Wang, Jingjing Wang, Hongwei Ouyang, Tingbo Liang*, Hongguang Xia*, Xiaoping Han*, and Guoji Guo*. A deep learning framework for in silico screening of anticancer drugs at the single-cell level. National Science Review, 2025, 12(2):nwae451. DOI: https://doi.org/10.1093/nsr/nwae451.
- SpeciesHuman
- Single cell388,646
- Major cell type86
- Cell lineage6
- Tissue6
- Cancer type7
- Screening compound~39,320