1. Framework
InTxDB is a comprehensive database dedicated to interactions involving bacteria type I-X secreted effectors and their host counterparts. It currently includes 1853 experimentally validated interaction pairs from 102 bacterial species. Detailed annotations are provided for protein sequences, functions, subcellular localizations, secretion signals, structures, and interaction sites. The database is regularly updated with new datasets.
1.1 Workflow
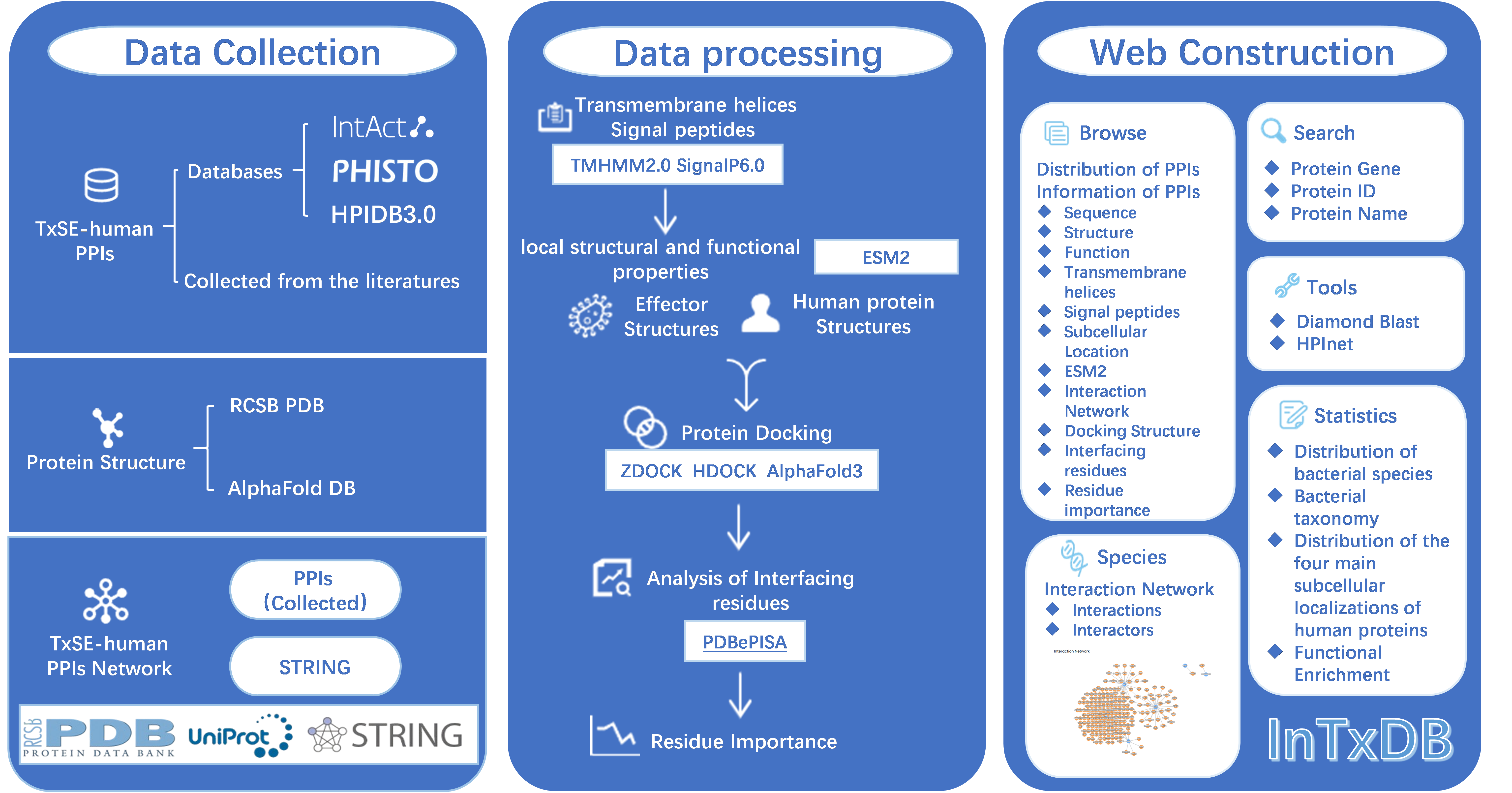 Fig. 1.1 Display the workflow.
Fig. 1.1 Display the workflow.
1.2 Data collection
We collected experimentally verified PPIs from five public databases (i.e. HPIDB 3.0[2], PHISTO [3], IntAct [1]) and recently published literature.
We collected experimentally verified Effectors from two public databases (i.e. SecReT4[4], TxSEdb [5]).
2. Search
Our database provides two main locations to perform searches: the Homepage and the Browse page. Both of these locations support queries for the following: bacterial protein name, bacteria species, UniProt ID, NCBI ID, human protein name, UniProt ID, gene name.
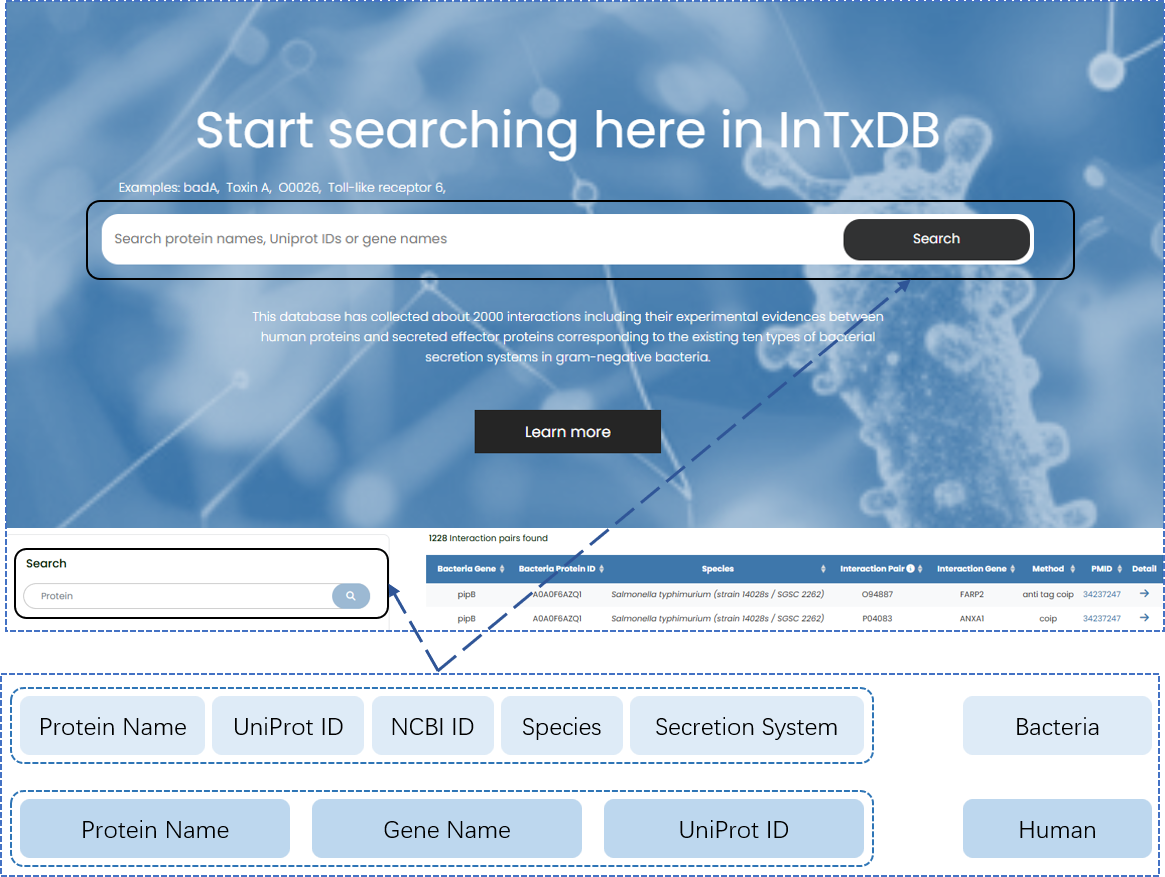 Fig. 2.1 Easy-to-access search interface that can accept multiple types of inputs.
Fig. 2.1 Easy-to-access search interface that can accept multiple types of inputs.
3. Browse
Statistical graphs on home page represent various secretion systems and species related to protein interaction. Users can simply click on any of these graphs to browse the interaction information. For a more personalized browsing experience, navigate to the 'Browse' page. Using a sidebar containing a variety of filters. You can select your desired secretion system and species from these filters.
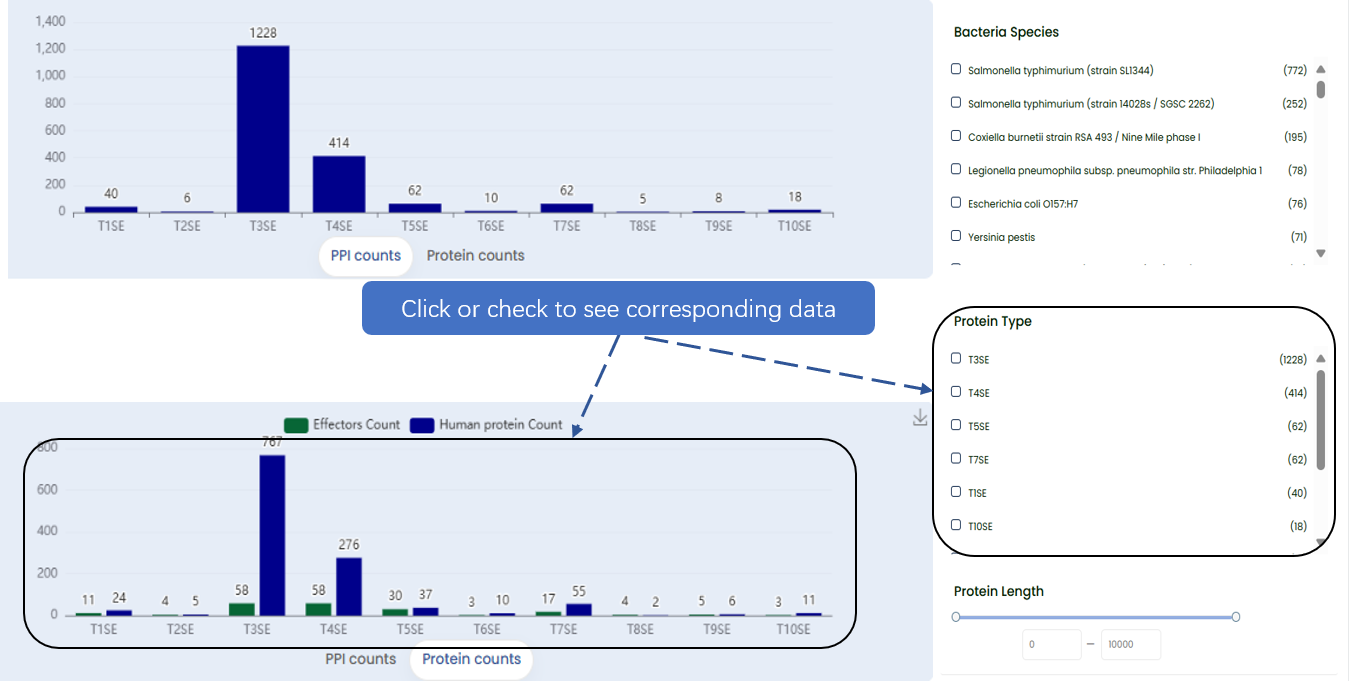 Fig. 3.1 Approaches to browse.
Fig. 3.1 Approaches to browse.
4. Species
Users can click on the navigation bar "Species", at which time the network diagram of the interaction between secreted proteins of each species and human proteins will appear. Meanwhile, Interactions between "Interactions" and "Interactors" will appear below. Users can freely choose to view each pair of interacting proteins of this species and the information of each protein
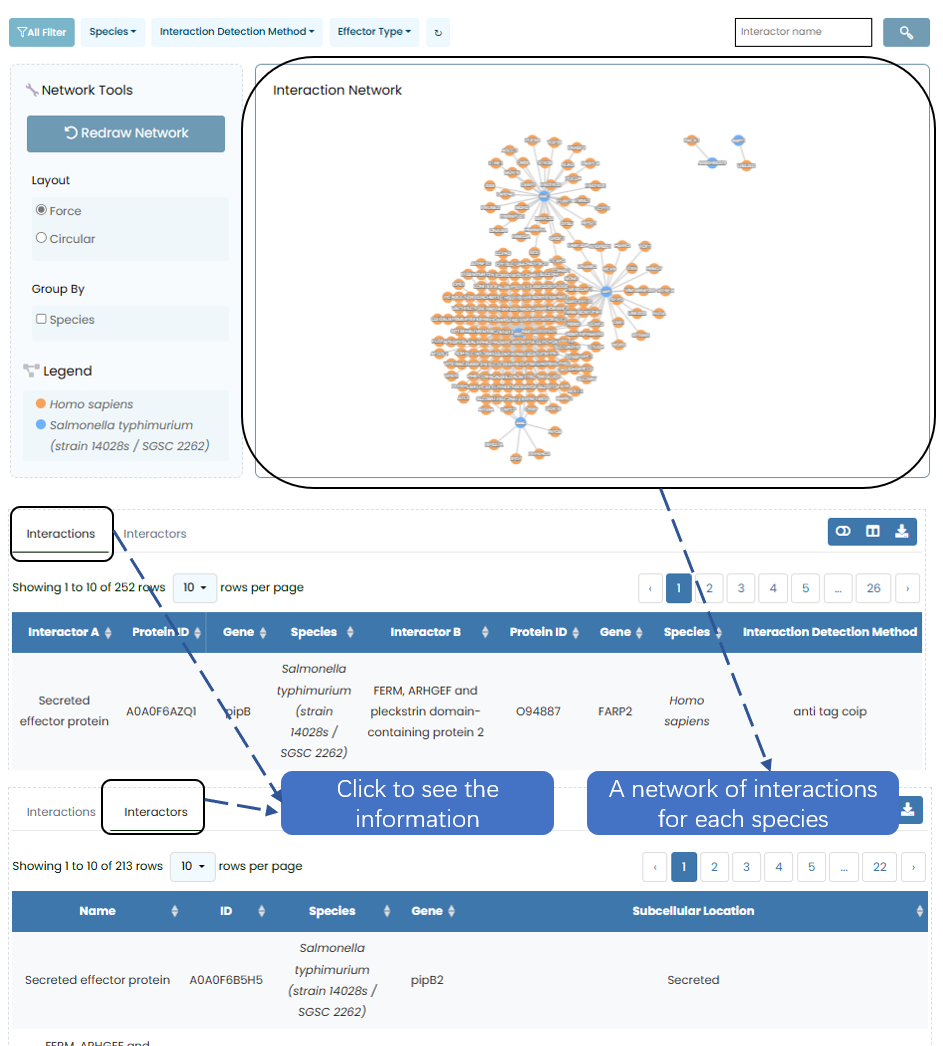 Fig. 3.1 Approaches to species.
Fig. 3.1 Approaches to species.
5. Interaction detail
In the 'browse' page, you can find details about the experimental validation methods used for the interactions and the literature sources for the information provided. To access more, simply find and click on the protein interaction you are interested in. Proceed to the detail page for an in-depth look of the interaction. Here, you will find comprehensive information about both interacting partners - the bacterium-secreted protein and the corresponding human protein. This includes their respective Protein-Protein Interaction (PPI) networks, experimental methods of verification, source literature, and docking information.
5.1 Protein information
The detailed characteristics of both interacting proteins: the bacterial protein and the corresponding human protein.
For your convenience, we have provided links to external databases which enables you to efficiently cross-reference and validate the information provided on our site. We employ the use of Mol* to provide you with a three-dimensional visualization of the protein structures. Instructions on how to use Mol* can be referred to their documentation.
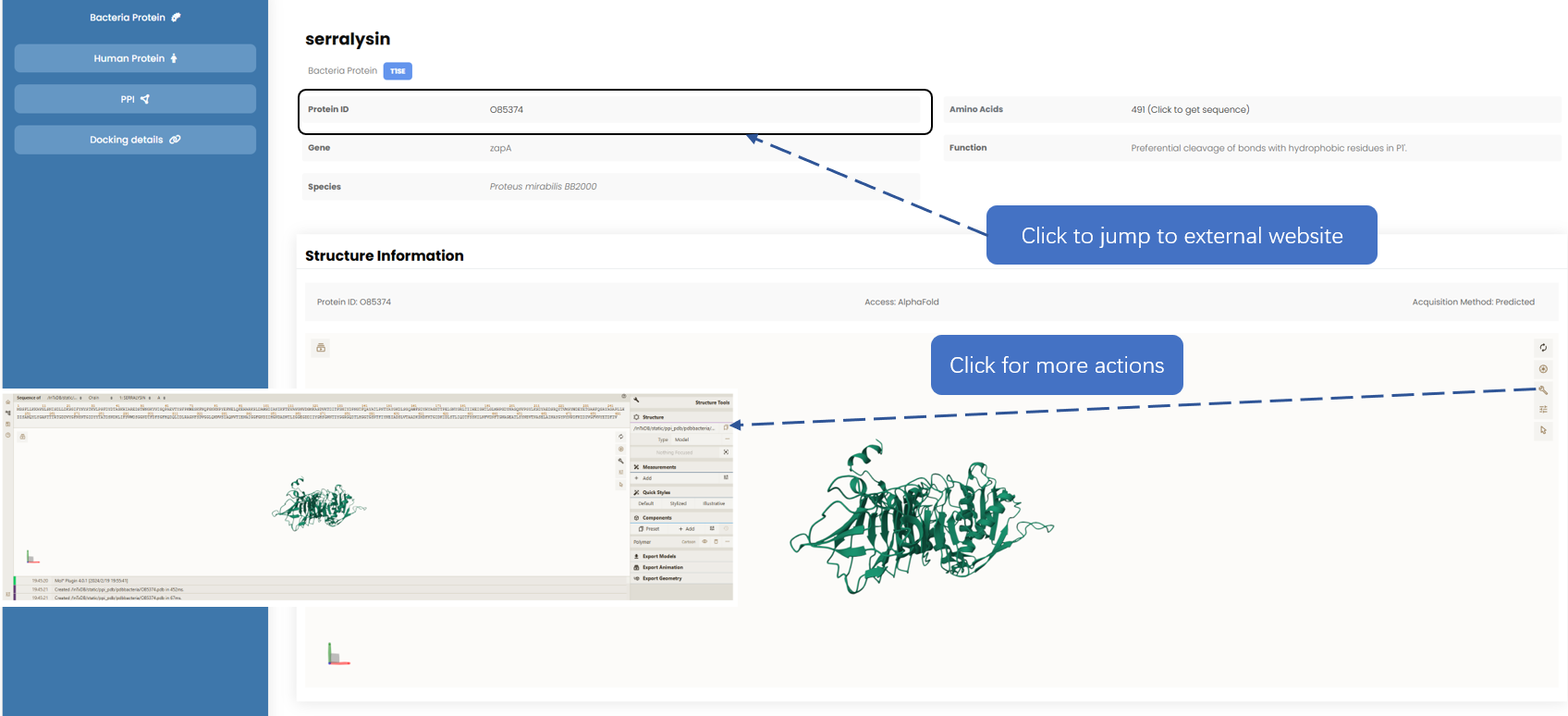 Fig. 5.1.1 Basic information.
Fig. 5.1.1 Basic information.
Our database provides a comprehensive visualization of the basic information of bacterial secretory proteins. We utilize TMHMM 2.0 to predict transmembrane helices and SignalP 6.0 to identify signal peptide sequences. And the results are presented in a visually accessible format.
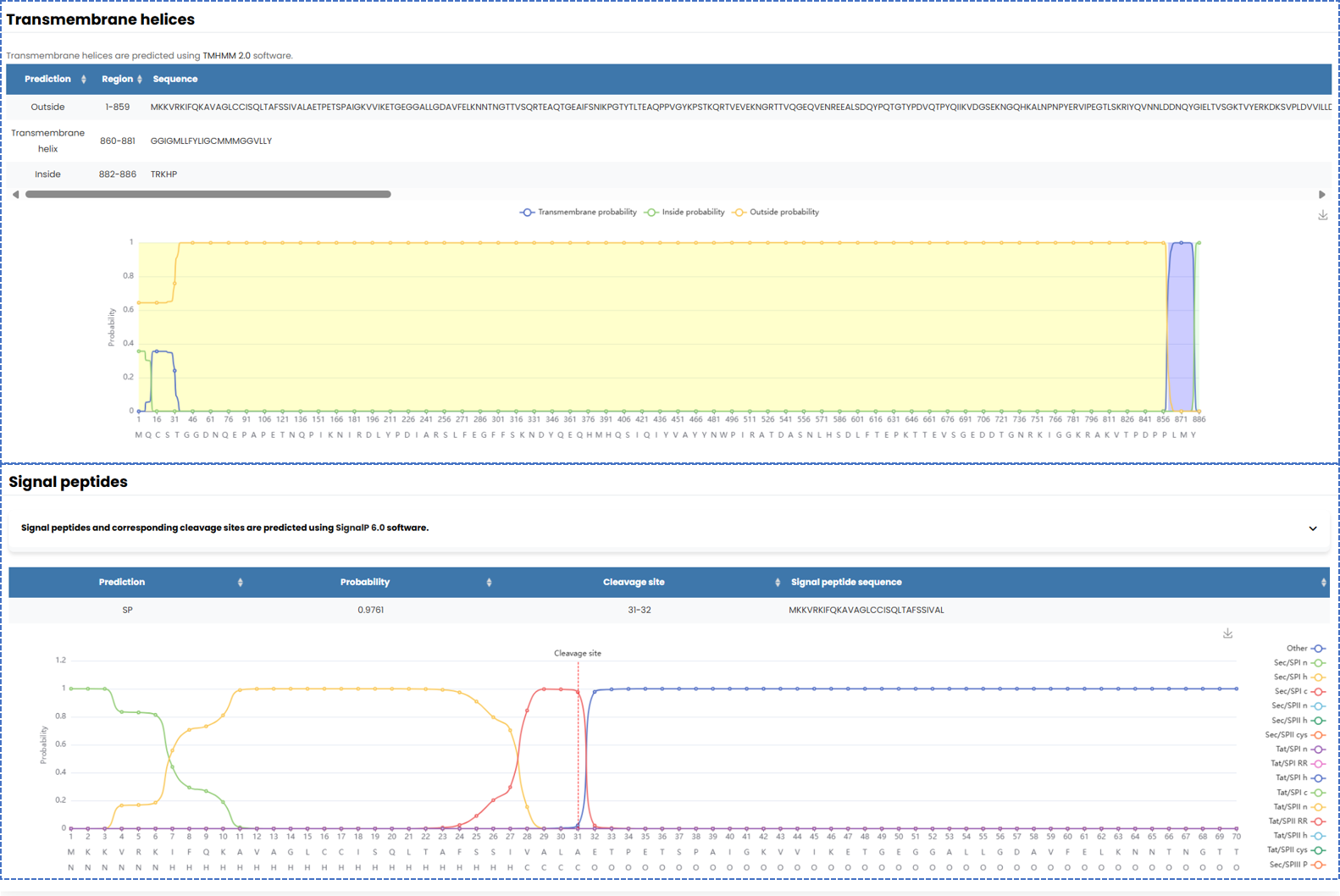 Fig. 5.1.2 Prediction and visulization of transmembrane helices and signal peptide sequences.
Fig. 5.1.2 Prediction and visulization of transmembrane helices and signal peptide sequences.
For bacterial proteins with unknown subcellular localization, we employ PSORTb v3.0 for prediction and display the results. Below is an overview of the software tools used:
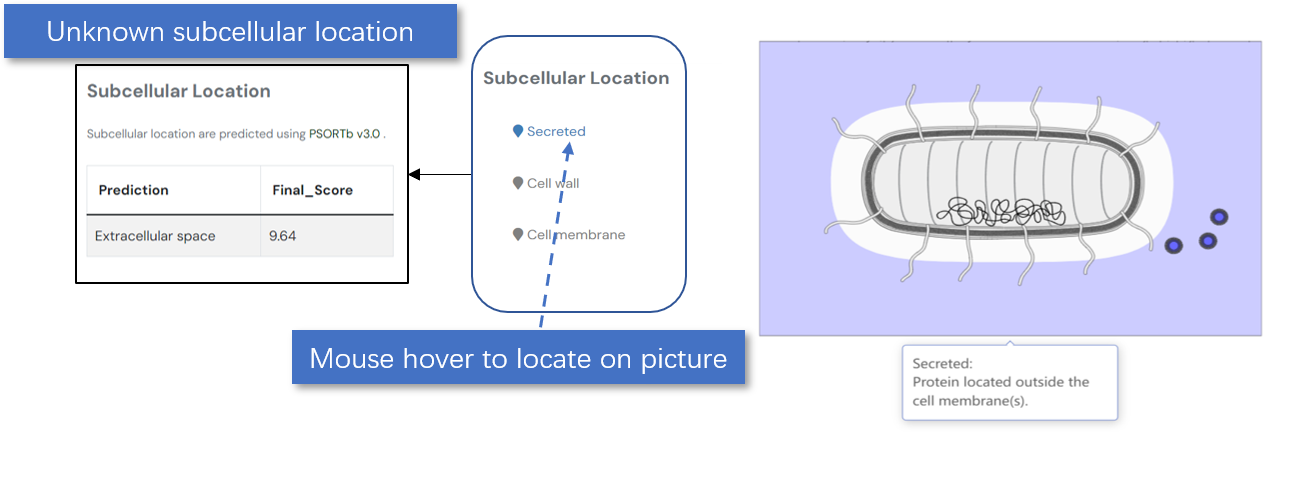 Fig. 5.1.3 Subcellular localization
Fig. 5.1.3 Subcellular localization
5.2 PPI
In addition to our own analyses, our database incorporates protein interaction networks from the STRING database. We have collected and displayed associated protein interaction network in every detail page.
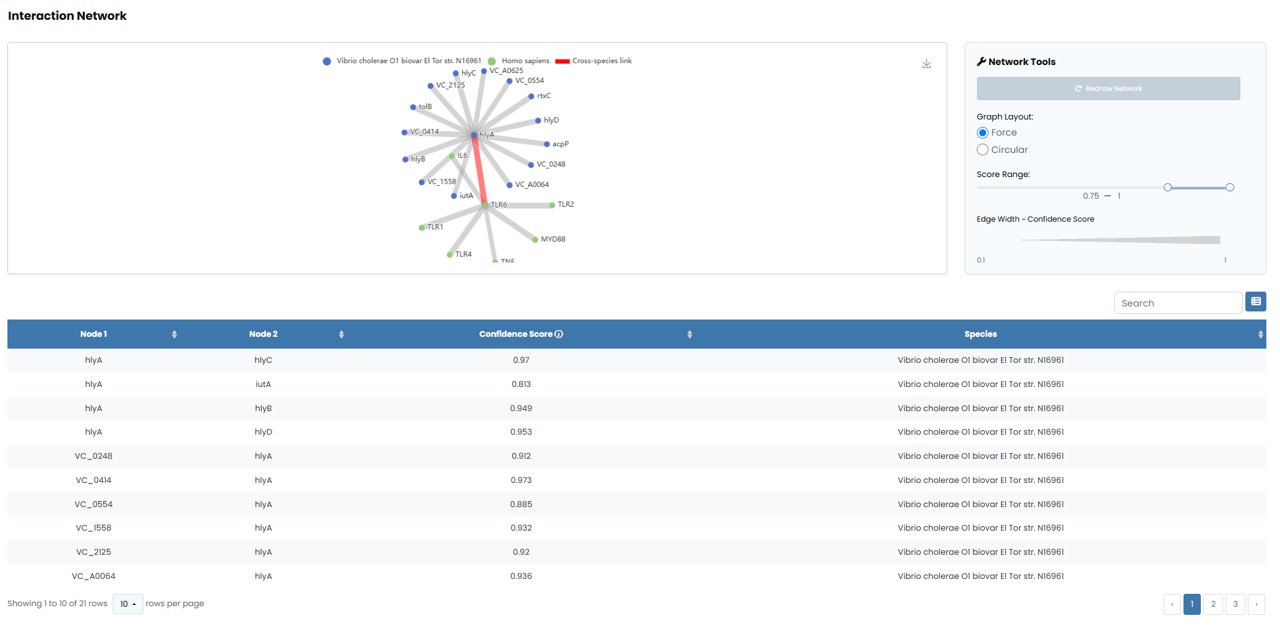 Fig. 5.2.1 PPI network information
Fig. 5.2.1 PPI network information
5.3 Docking prediction
To provide a comprehensive understanding of the interactions between bacterial proteins and human proteins, We employ computational methods including AlphaFold3 , ZDOCK and HDOCK to predict the docking sites of each protein interaction pair. Users can click on each column to display the structure of each prediction tool
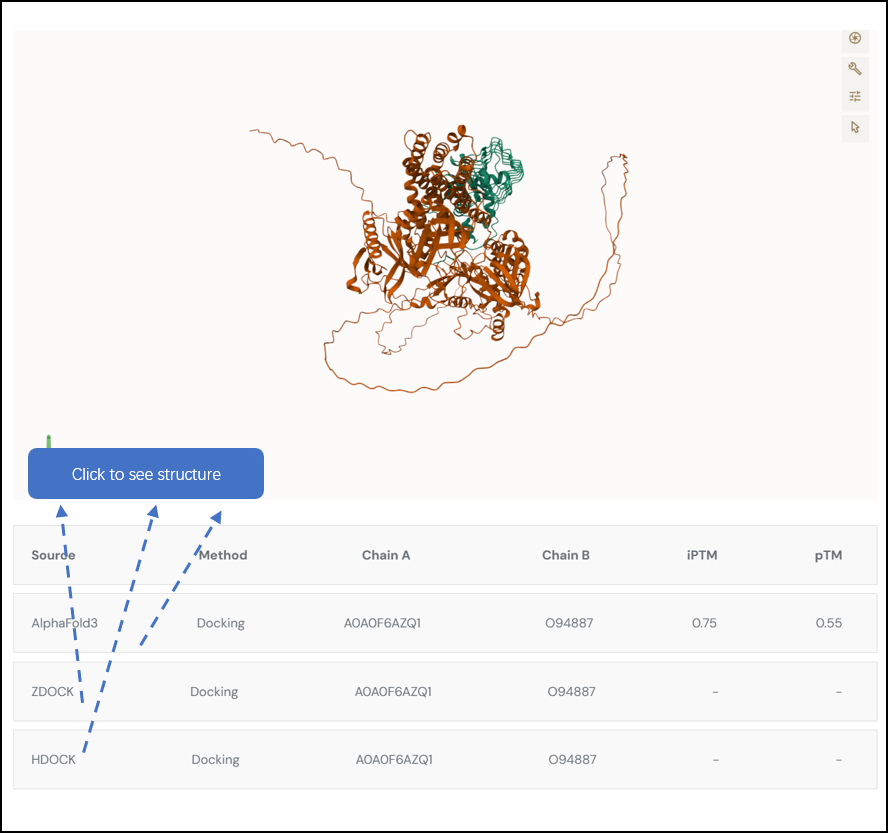 Fig. 5.3.1 Docking prediction
Fig. 5.3.1 Docking prediction
Below is the data obtained by the three docking methods after PDBePISA analysis (the data is accurate to two decimal places), and the line chart of the importance of each residue analyzed by the three docking methods and our method, HPInet
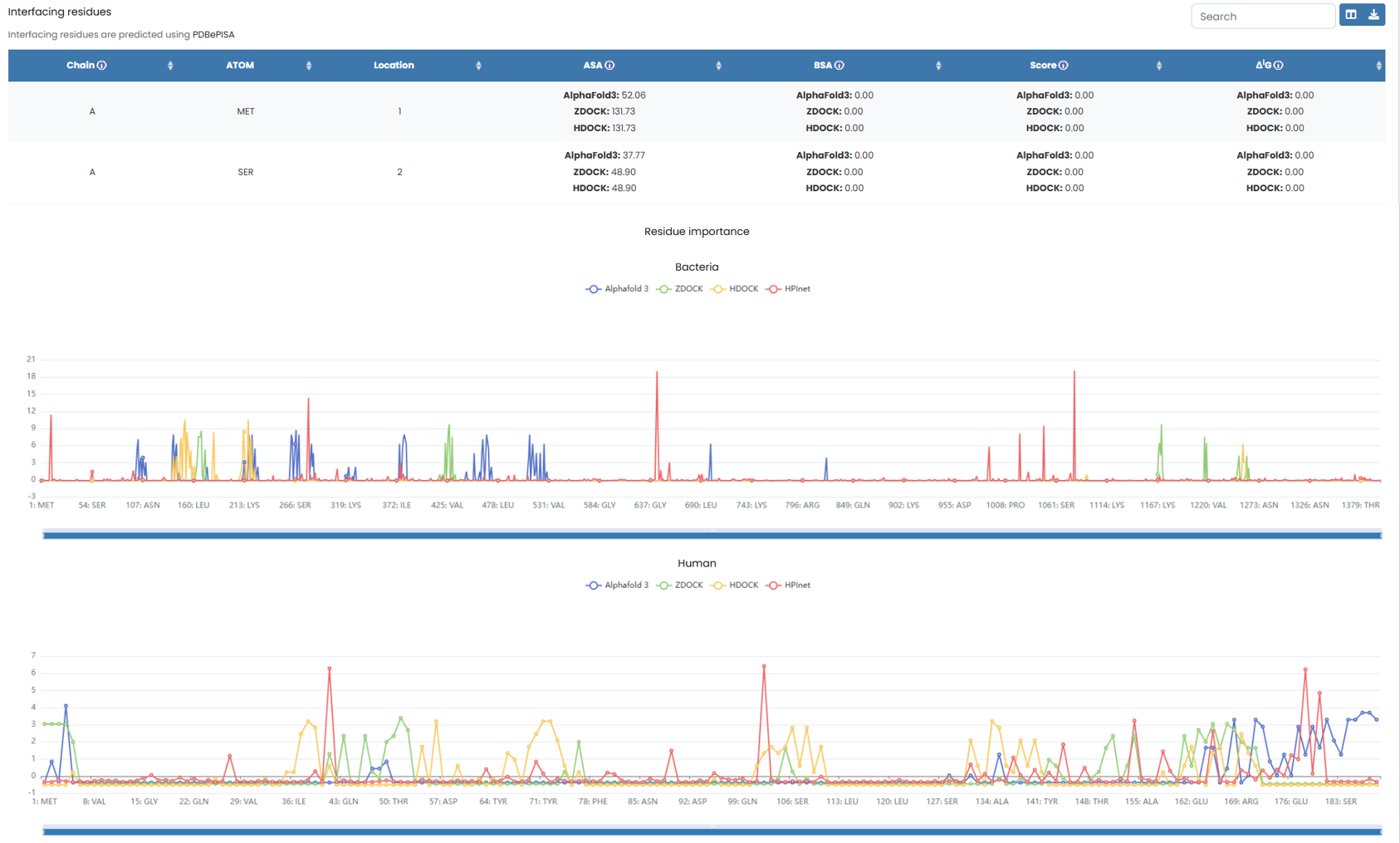 Fig. 5.3.2 PDBePISA analysis results and importance analysis of each residue.
Fig. 5.3.2 PDBePISA analysis results and importance analysis of each residue.
6. Tools
The following two tools can be used to analyze the interaction between other similar proteins and the interaction between unknown proteins.
Blast
Diamond (DIAMOND) BLAST is an efficient sequence alignment tool, through which you can obtain the results of the sequence after blast and the network graph structure obtained after blast
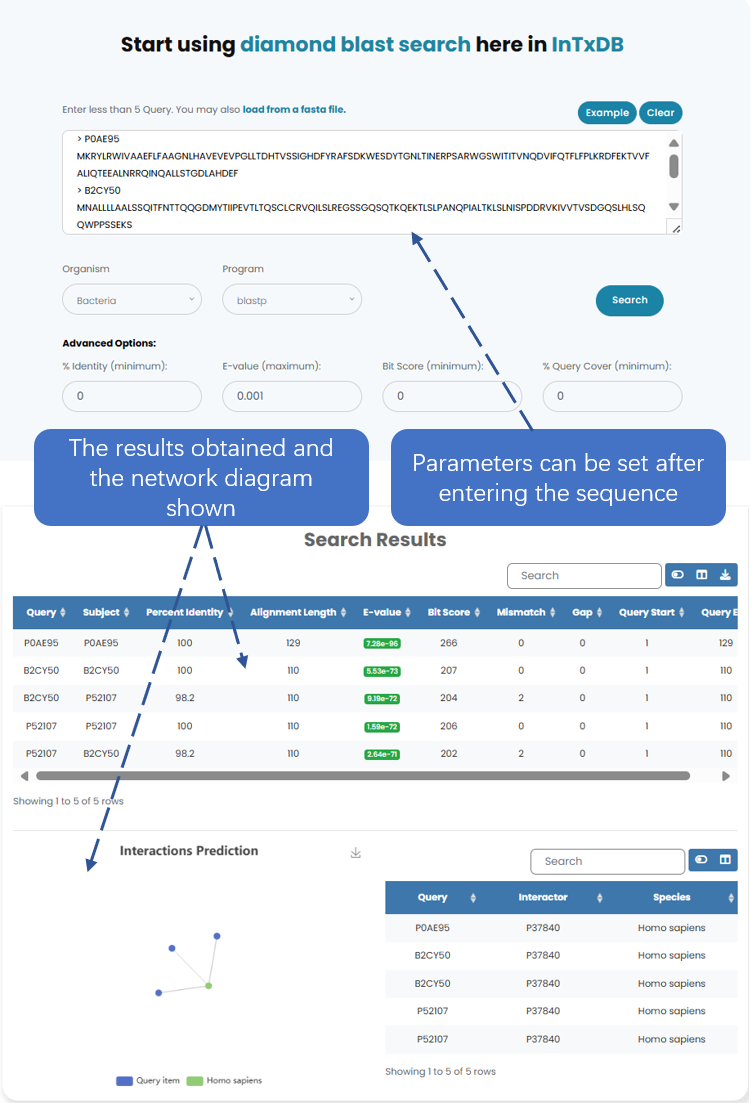 Fig. 6.1 Display the result after blast input.
Fig. 6.1 Display the result after blast input.
5.2 Prediction
HPInet is a Transformer-based predictor for rapid prediction of distant substrate secretion protein-host protein interactions. HPInet is first deep learning model specifically designed for interpretable predictions of secreted effectors and host proteins interactions, allowing for the rapid identification of potential interaction sites.
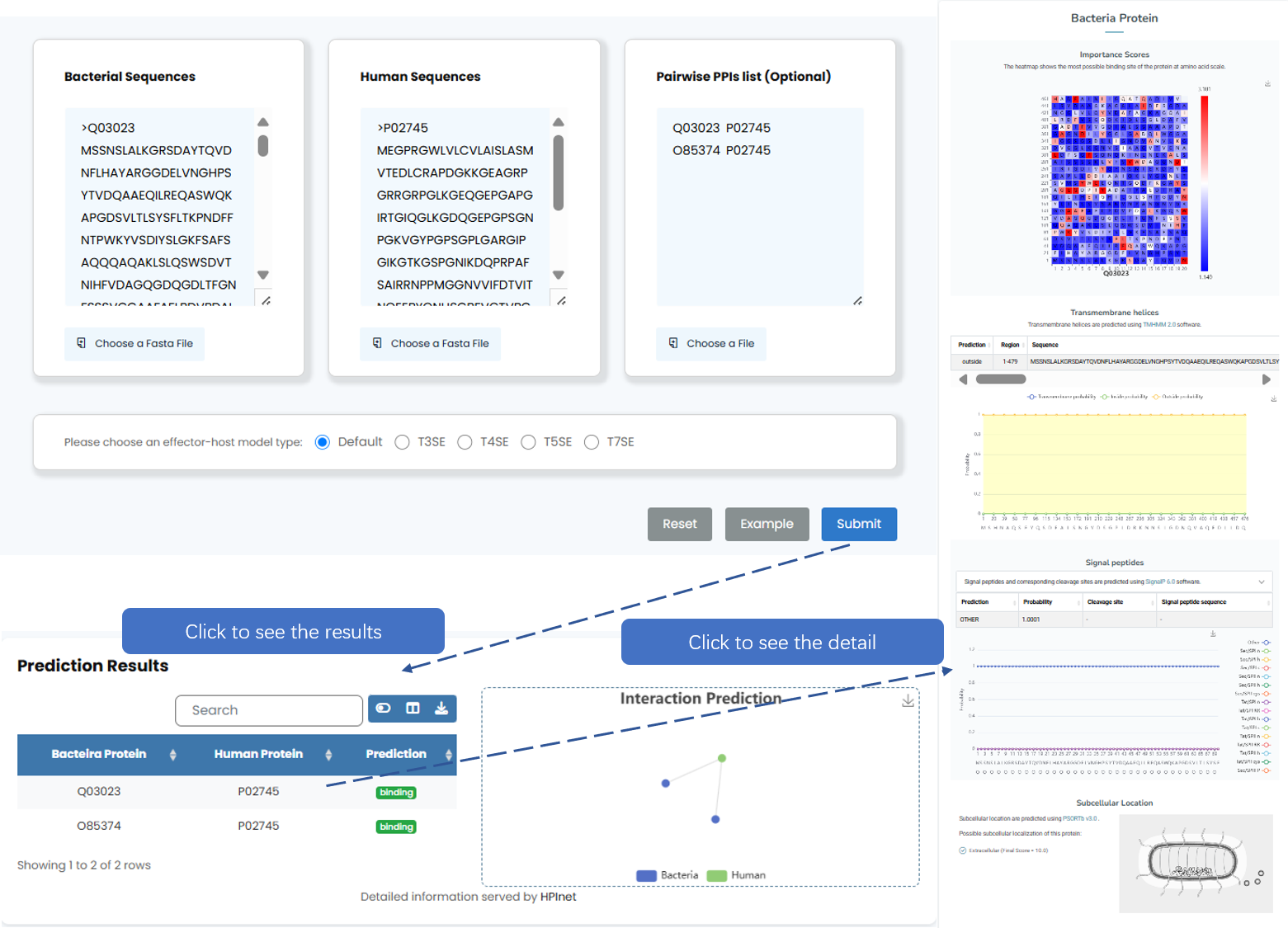 Fig. 6.2 PPI network information
Fig. 6.2 PPI network information
7. Statistics
The page has three diagrams showing the number of species for each secretion type, taxonomic statistics for all species included in the data (click to access the relevant browse page), and subcellular localization of the human proteins contained in each secretion type
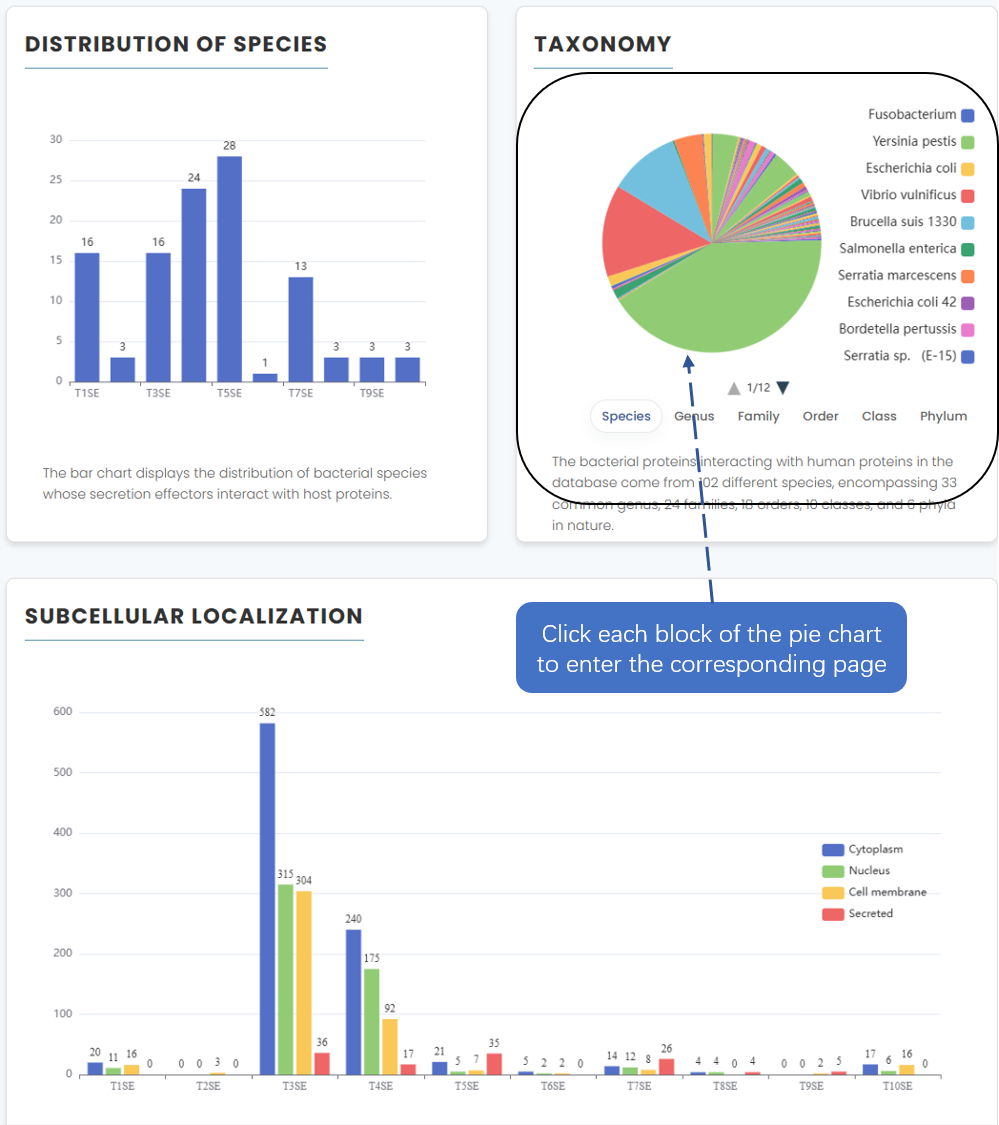 Fig. Statistics page display.
Fig. Statistics page display.
8. Contact
We value your insights and are eager to address any questions or comments you may have. You can find our contact details here. We look forward to hearing from you and engaging in meaningful conversations.
Ming Chen's lab, College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, China
Ø Telephone: +86 (0)571-88206612
Ø Fax: +86 (0)571-88206612
Ø E-mail: Ming Chen (mchen@zju.edu.cn); Yueming Hu (huym@zju.edu.cn); Liya Liu (3190103446@zju.edu.cn); Yanyan Zhu (22207031@zju.edu.cn)
9. Reference
1. Del Toro N, Shrivastava A, Ragueneau E, et al. The IntAct database: efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022 Jan 7;50(D1):D648-D653. doi: 10.1093/nar/gkab1006. PMID: 34761267.
2. Ammari MG, Gresham CR, McCarthy FM, et al. HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford). 2016 Jul 3:2016:baw103. doi: 10.1093/database/baw103. PMID: 27374121.
3. Durmuş Tekir S, Çakir T, Ardiç E, et al. PHISTO: pathogen-host interaction search tool. Bioinformatics. 2013 May 15;29(10):1357-8. doi: 10.1093/bioinformatics/btt137. PMID: 23515528.
4. Bi D, Liu L, Tai C, et al. SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 2013 Jan;41(Database issue):D660-5. doi: 10.1093/nar/gks1248. PMID: 23193298.
5. Hui X, Chen Z, Zhang J, et al. Computational prediction of secreted proteins in gram-negative bacteria. Comput Struct Biotechnol J. 2021 Mar 22:19:1806-1828. doi: 10.1016/j.csbj.2021.03.019. PMID: 33897982.
10. Data Submission
We welcome contributions from the research community to continuously expand and improve InTxDB. If you have newly identified or experimentally validated protein-protein interactions between Gram-negative bacterial secreted effectors and human host proteins, you can submit them for inclusion in future database updates. To submit your data, please email us at mchen@zju.edu.cn with the following information:
- 1. Effector protein name and UniProt ID (or sequence)
- 2. Human target protein name and UniProt ID (or sequence)
- 3. Bacterial species and secretion system type
- 4. Experimental validation method(s) used (e.g., Co-IP, Y2H, MS)
- 5. Literature reference (PubMed ID or DOI)
- 6. (Optional) Structural information or predicted models
All submitted entries will undergo manual curation and verification before being integrated into the database. We appreciate your support in building a more comprehensive resource for the host-pathogen interaction research community.
11. Citation
If you use InTxDB in your research, please cite the following publication:
-
Zhu Y, Liu L, Hu Y, Li S, Liu E, Hu Y, Zhang S, Chao H, Fang Q, Huan Y, Chen M. InTxDB: A curated database of experimentally validated interactions between Gram-negative bacterial effectors and human host proteins. Database, 2025.
Please include this citation in any publications, presentations, or derivative works that utilize data, tools, or visualizations from InTxDB.