Help document
1. Introduction of PSGDb
Plants growth and productivity are greatly
affected by both biotic and abiotic stresses. However, in order to
live and propagate on the earth, plants have developed a series of
fine mechanisms for responding to environmental changes, which
have been established during their long-period evolution and
domestication. Researchers have found that there are many stress
resistance genes existed in plants and they play important roles
in the resistance to the environmental adversity. So the deep
research about these genes seems quite essential. For this
purpose, a database resource named PSGDb (Plant Stress Gene
Database) was developed to systematically document and integrate
these various stress data and its genetic relations, which could
further help us to gain a global insight into plant stress genes
and how they function with each other. Besides, several useful
analysis tools were also designed to facilitate us to find more
potential stress genes. Database URL:
http://bis.zju.edu.cn/PSGDb/index.html
2. Database Design and Construction
PSGDb integrates several public databases,
including GenBank , UniProt , PubMed , Gene Ontology (GO), KEGG ,
Tair and UniGene. GenBank was searched by using E-utilities with
the keywords of “stress”, “resistance”, “response to” and more.
During this process the organism section was limited to plants.
Several perl scripts were used to download the data from GenBank,
remove those replaced or discontinued entries collected and
extract useful information from thousands of gene data files.
GenBank will provide the basic information about the plant stress
genes. The protein information is also supplied if there is a
corresponding protein entry in UniProt and the related literatures
are integrated from PubMed. Besides, GO and KEGG of the stress
genes are also available. Tair is direct linked as an important
complement to these entries specific to Arabidopsis thaliana.
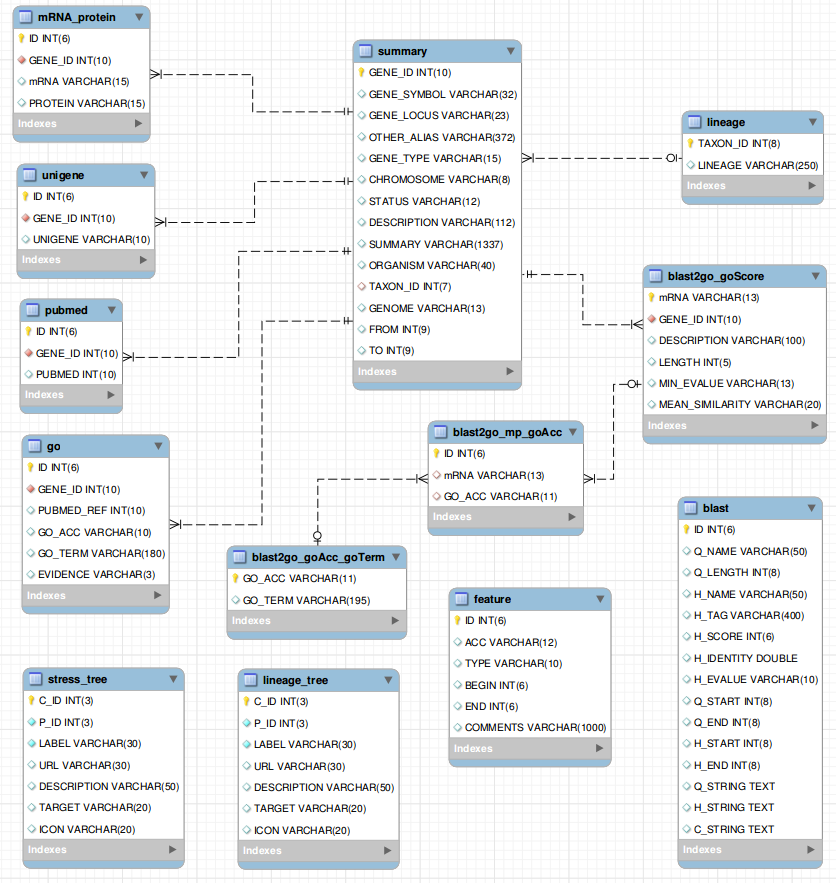
3. Main Techniques
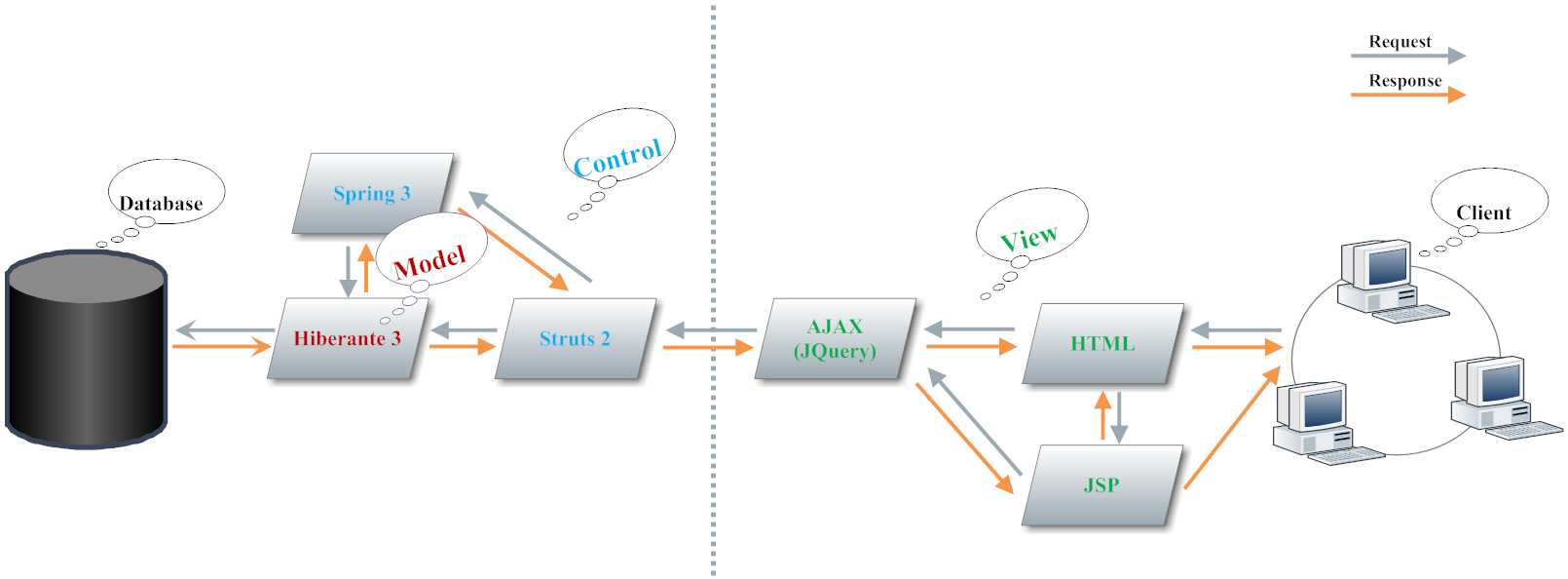
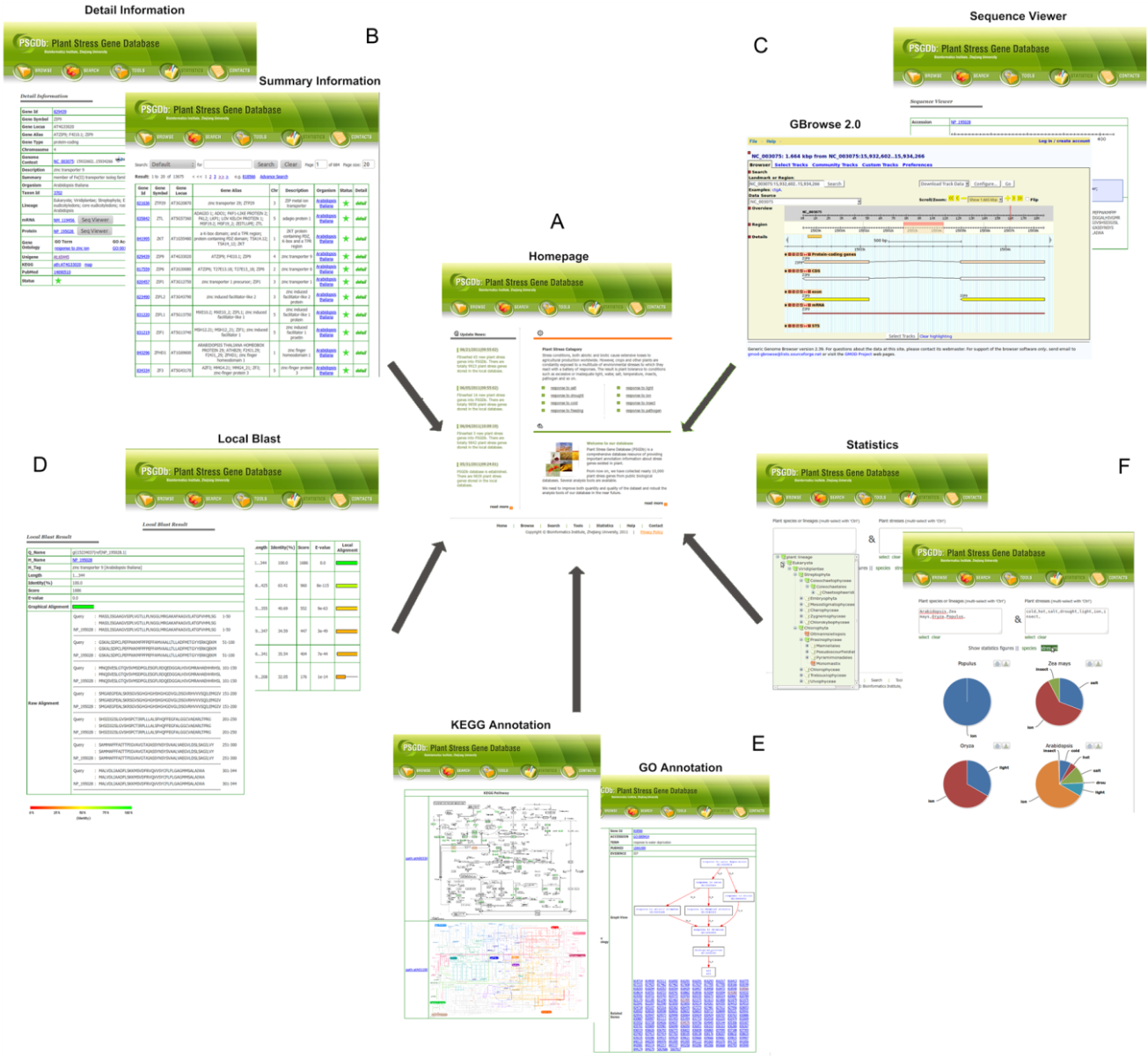
4. Application of PSGDb
PSGDb is a quite comprehensive database
resource of plant stress genes not only maintains a lot of plant
species, but also involves almost complete plant stresses, from
which a full-scale insight of the functions of these plant stress
genes could be gained. Besides, it also developed five useful
analysis tools by using the database, they are: Advanced
Search(B), Gbrowse2.0 & Sequence Viewer(C), Local Blast(D),
GO/KEGG Annotation(E) and Statistics(F).
Database Query
By searching PSGDb (basic or advanced search), you will go to the Summary Information page. The summary information of the genes includes Gene ID, Gene Name, Protein Name, Locus Tag, Chromosome, GI, Genome context, Organism, Taxon Id and Status. The "detail" icon in the summary information page will allow you to visit the detail information page of the specific gene. The detail information of a gene includes all of the information shown in the summary information page as well as other important annotation information. In this page, you could use GBrowse2.0 to view the Genome context of the gene. The corresponding NCBI entries could be visited by clicking the NCBI Nucleotide ID or Protein Accession ID and the SeqViewer page of that gene could be shown by clicking the magnifier icon. The hyperlink of genbank/fasta/gff is used to download the specific format file of a gene. There are some references to other biology databases, including Swissprot, Unigene, KEGG and Gene Ontology.
Advanced Search
The Advanced Search is designed for a more sophisticated and efficient search method by using the boolean words (AND, OR, NOT). It allows users to search almost every aspect of the plant stress genes, like gene id, gene name, protein name and so on. If the users input the word correctly in these search boxes, it will automatically prompt the more likely keywords for users to choose directly.
Gbrowse2.0 & Sequence Viewer
As we know, GBrowse2.0 is a genome viewer and is GMOD’s most popular component. To view the sequences related to plant stress genes by GBrowse2.0 that displays a more user-friendly sequence viewer interface. Furthermore, an in-house developed sequence viewer (SeqViewer) functioned as a complement of GBrowse2.0 is available. Figure 1D show the genomic sequence of NC_000927:1..200799. SeqViewer contains two parts: the Sequence Feature part and the Sequence part. The former shows the features of CDSs, Nucleotides/Proteins, Regions, Sites and sources while the latter displays the sequences with which you could do local blast.
Local Blast
Local Blast Alignment allows users to blast against PSGDb by using the entries in PSGDb or their own sequences files. The file’s format should be Fasta or flat text. Users could use the default parameters or set their preferred values through the optional Advanced Settings. Figure 1C shows the blast result of protein NP_973396.1 which includes the information of Q_Name, H_Name, H_tag, Length, Identity (%), Score, E-value and Local Alignment. In the Local Alignment field, the greener of the bar icon’s colour, the closer to 100 of the Identity while the redder of the bar icon’s colour, the closer to 0 of the Identity. When one of the blast alignment bars is chosen, the specific sequences alignment will be shown.
Database Query
By searching PSGDb (basic or advanced search), you will go to the Summary Information page. The summary information of the genes includes Gene ID, Gene Name, Protein Name, Locus Tag, Chromosome, GI, Genome context, Organism, Taxon Id and Status. The "detail" icon in the summary information page will allow you to visit the detail information page of the specific gene. The detail information of a gene includes all of the information shown in the summary information page as well as other important annotation information. In this page, you could use GBrowse2.0 to view the Genome context of the gene. The corresponding NCBI entries could be visited by clicking the NCBI Nucleotide ID or Protein Accession ID and the SeqViewer page of that gene could be shown by clicking the magnifier icon. The hyperlink of genbank/fasta/gff is used to download the specific format file of a gene. There are some references to other biology databases, including Swissprot, Unigene, KEGG and Gene Ontology.
Advanced Search
The Advanced Search is designed for a more sophisticated and efficient search method by using the boolean words (AND, OR, NOT). It allows users to search almost every aspect of the plant stress genes, like gene id, gene name, protein name and so on. If the users input the word correctly in these search boxes, it will automatically prompt the more likely keywords for users to choose directly.
Gbrowse2.0 & Sequence Viewer
As we know, GBrowse2.0 is a genome viewer and is GMOD’s most popular component. To view the sequences related to plant stress genes by GBrowse2.0 that displays a more user-friendly sequence viewer interface. Furthermore, an in-house developed sequence viewer (SeqViewer) functioned as a complement of GBrowse2.0 is available. Figure 1D show the genomic sequence of NC_000927:1..200799. SeqViewer contains two parts: the Sequence Feature part and the Sequence part. The former shows the features of CDSs, Nucleotides/Proteins, Regions, Sites and sources while the latter displays the sequences with which you could do local blast.
Local Blast
Local Blast Alignment allows users to blast against PSGDb by using the entries in PSGDb or their own sequences files. The file’s format should be Fasta or flat text. Users could use the default parameters or set their preferred values through the optional Advanced Settings. Figure 1C shows the blast result of protein NP_973396.1 which includes the information of Q_Name, H_Name, H_tag, Length, Identity (%), Score, E-value and Local Alignment. In the Local Alignment field, the greener of the bar icon’s colour, the closer to 100 of the Identity while the redder of the bar icon’s colour, the closer to 0 of the Identity. When one of the blast alignment bars is chosen, the specific sequences alignment will be shown.
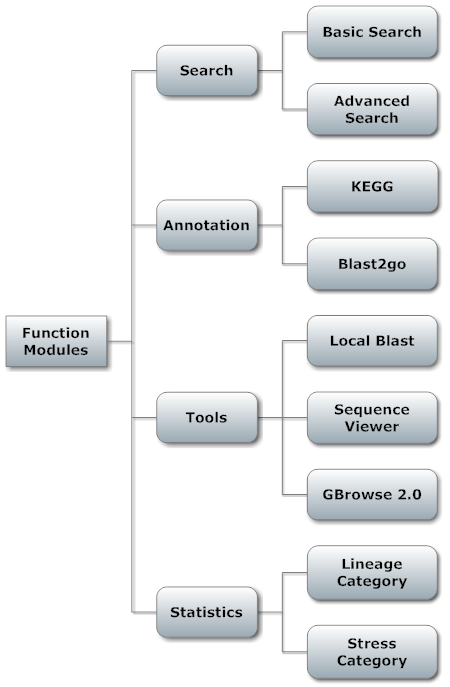
Function Modules of PSGDb
5. References
1. Puijalon, S., et al., Clonal plasticity of aquatic plant
species submitted to mechanical stress: escape versus resistance
strategy.
Ann Bot, 2008.
102(6): p. 989-996.
2. Meguro, N., D. Saisho, and M. Nakazono, [Role of plant mitochondria in avoidance of oxidative stress]. Tanpakushitsu Kakusan Koso, 2005. 50(14 Suppl): p. 1879-1882.
3. Wan, X., et al., Increased tolerance to oxidative stress in transgenic tobacco expressing a wheat oxalate oxidase gene via induction of antioxidant enzymes is mediated by H2O2. Physiol Plant, 2009. 136(1): p. 30-44.
4. Handa, S., et al., Solutes contributing to osmotic adjustment in cultured plant cells adapted to water stress. Plant Physiol, 1983. 73(3): p. 834-843.
5. Popko, J., et al., The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol (Stuttg). 12(2): p. 242-258.
6. Engelberth, M.J. and J. Engelberth, Monitoring plant hormones during stress responses. J Vis Exp, 2009(28).
7. Kurepa, J., et al., Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav, 2009. 4(10): p. 924-927.
8. Sanseverino, W., et al., PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res, 2010. 38(Database issue): p. D814-21.
9. Vleeshouwers, V.G., et al., SolRgene: an online database to explore disease resistance genes in tuber-bearing Solanum species. BMC Plant Biol, 2011. 11: p. 116.
10. Hammami, R., et al., PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res, 2009. 37(Database issue): p. D963-8.
11. Li, D., et al., An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics, 2009. 10: p. 517.
12. Shameer, K., et al., STIFDB-Arabidopsis Stress Responsive Transcription Factor DataBase. Int J Plant Genomics, 2009. 2009: p. 583429.
13. Benson, D.A., et al., GenBank. Nucleic Acids Res, 2009. 37(Database issue): p. D26-31.
14. Apweiler, R., et al., UniProt: the Universal Protein knowledgebase. Nucleic Acids Res, 2004. 32(Database issue): p. D115-119.
15. McEntyre, J. and D. Lipman, PubMed: bridging the information gap. CMAJ, 2001. 164(9): p. 1317-1319.
16. Ashburner, M., et al., Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 2000. 25(1): p. 25-29.
17. Kanehisa, M. and S. Goto, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 2000. 28(1): p. 27-30.
18. Calcitonin intranasal--unigene: Salcatonin intranasal--unigene. Drugs R D, 2004. 5(2): p. 90-93.
19. Conesa, A., et al., Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics , 2005. 21(18): p. 3674-3676.
20. Healy, M.D., Using BLAST for performing sequence alignment. Curr Protoc Hum Genet, 2007. Chapter 6: p. Unit 6 8.
21. Stein, L.D., et al., The generic genome browser: a building block for a model organism system database. Genome Res, 2002. 12(10): p. 1599-1610.
2. Meguro, N., D. Saisho, and M. Nakazono, [Role of plant mitochondria in avoidance of oxidative stress]. Tanpakushitsu Kakusan Koso, 2005. 50(14 Suppl): p. 1879-1882.
3. Wan, X., et al., Increased tolerance to oxidative stress in transgenic tobacco expressing a wheat oxalate oxidase gene via induction of antioxidant enzymes is mediated by H2O2. Physiol Plant, 2009. 136(1): p. 30-44.
4. Handa, S., et al., Solutes contributing to osmotic adjustment in cultured plant cells adapted to water stress. Plant Physiol, 1983. 73(3): p. 834-843.
5. Popko, J., et al., The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol (Stuttg). 12(2): p. 242-258.
6. Engelberth, M.J. and J. Engelberth, Monitoring plant hormones during stress responses. J Vis Exp, 2009(28).
7. Kurepa, J., et al., Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav, 2009. 4(10): p. 924-927.
8. Sanseverino, W., et al., PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res, 2010. 38(Database issue): p. D814-21.
9. Vleeshouwers, V.G., et al., SolRgene: an online database to explore disease resistance genes in tuber-bearing Solanum species. BMC Plant Biol, 2011. 11: p. 116.
10. Hammami, R., et al., PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res, 2009. 37(Database issue): p. D963-8.
11. Li, D., et al., An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics, 2009. 10: p. 517.
12. Shameer, K., et al., STIFDB-Arabidopsis Stress Responsive Transcription Factor DataBase. Int J Plant Genomics, 2009. 2009: p. 583429.
13. Benson, D.A., et al., GenBank. Nucleic Acids Res, 2009. 37(Database issue): p. D26-31.
14. Apweiler, R., et al., UniProt: the Universal Protein knowledgebase. Nucleic Acids Res, 2004. 32(Database issue): p. D115-119.
15. McEntyre, J. and D. Lipman, PubMed: bridging the information gap. CMAJ, 2001. 164(9): p. 1317-1319.
16. Ashburner, M., et al., Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 2000. 25(1): p. 25-29.
17. Kanehisa, M. and S. Goto, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 2000. 28(1): p. 27-30.
18. Calcitonin intranasal--unigene: Salcatonin intranasal--unigene. Drugs R D, 2004. 5(2): p. 90-93.
19. Conesa, A., et al., Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics , 2005. 21(18): p. 3674-3676.
20. Healy, M.D., Using BLAST for performing sequence alignment. Curr Protoc Hum Genet, 2007. Chapter 6: p. Unit 6 8.
21. Stein, L.D., et al., The generic genome browser: a building block for a model organism system database. Genome Res, 2002. 12(10): p. 1599-1610.