| Home | Browser | Searcher | Viewer | Statistics | Download | Documentation |
|
Document:
Table of Contents
1 Data Collection & Processing 
1.1 Gene Information 
All the gene annotation information for the 70 plant species can be obtained from the URLs provided at the Data Sources tab of this page.
1.2 GO Annotaion 
GO annotation information was download from the Gene Ontology database (http://www.geneontology.org/; Release 2010-08-01) [24].
1.3 Small RNA Data Sets 
♦ Small RNA(sRNA) high-throughput sequencing data sets of each species were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [27]. All the sRNA data sets retrieved for this study were summarized in the Statistics page → Small RNA Datasets tab.
♦ SRNA sequences containing incomplete information (such as containing "N") and with length less than 18 or more than 28 were removed for further analysis. For each data set, the filtered sRNA sequences were mapped to all the gene models of the related plant species. All mapping steps were performed using the Bowtie algorithm [33] allowing no mismatches. Besides, for comparison, the normalized abundance of sRNAs from each data set was calculated as RPMs (reads per million), which divided the read number of each sRNA by the total reads from this data set, and multiplied by 106.
1.4 Other Information 
The transcription factor (TF) information for each species (if available) was retrieved from two TF databases: PlantTFDB (Plant Transcription Factor Database; http://planttfdb.cbi.pku.edu.cn/index.php) and PlnTFDB (The Plant Transcription Factor Database; http://plntfdb.bio.uni-potsdam.de/v3.0/)
2 Browser 
Browser is a quick approach to access your interested information. You can browse your interested species by either "Browse by Species" or "Browse by Classification".
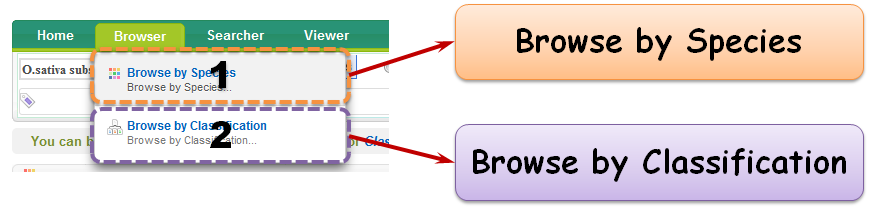
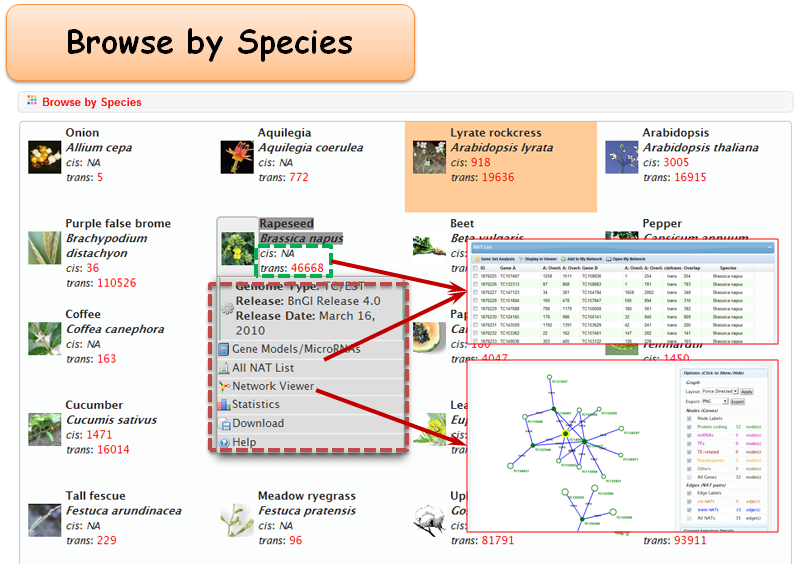
2.1 Browse by Species 
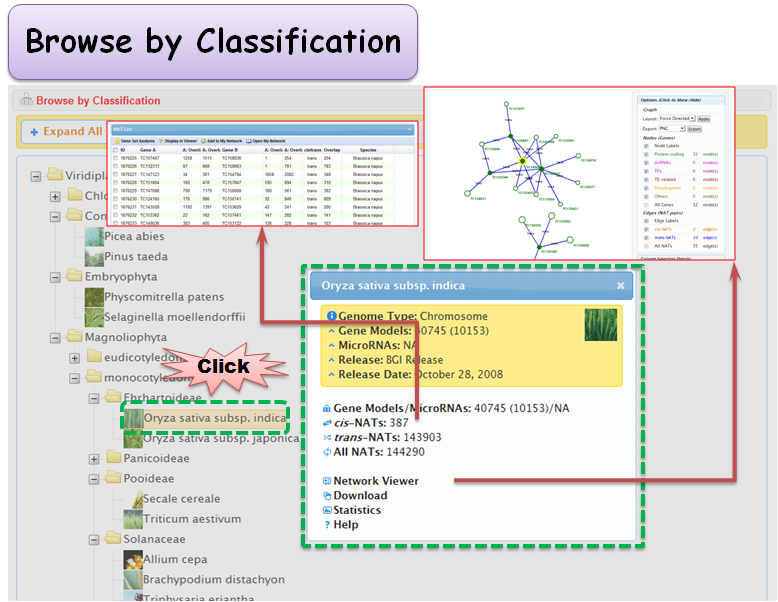
2.2 Browse by Classification 
3 Searcher 
Search can be performed by "Quick Searcher", "Simple Searcher", "Batched Searcher", "Advanced Searcher" or "BLAST Searcher".
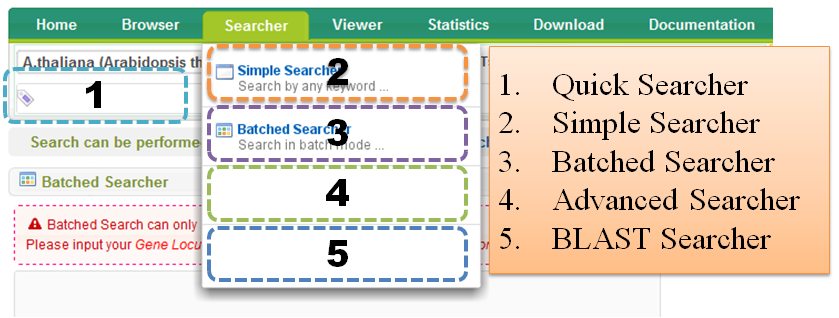
3.1 Quick Searcher 
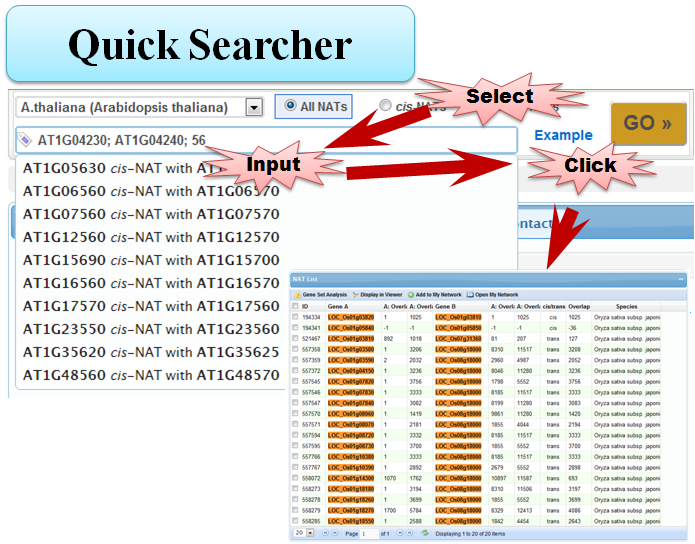
3.2 Simple Searcher 

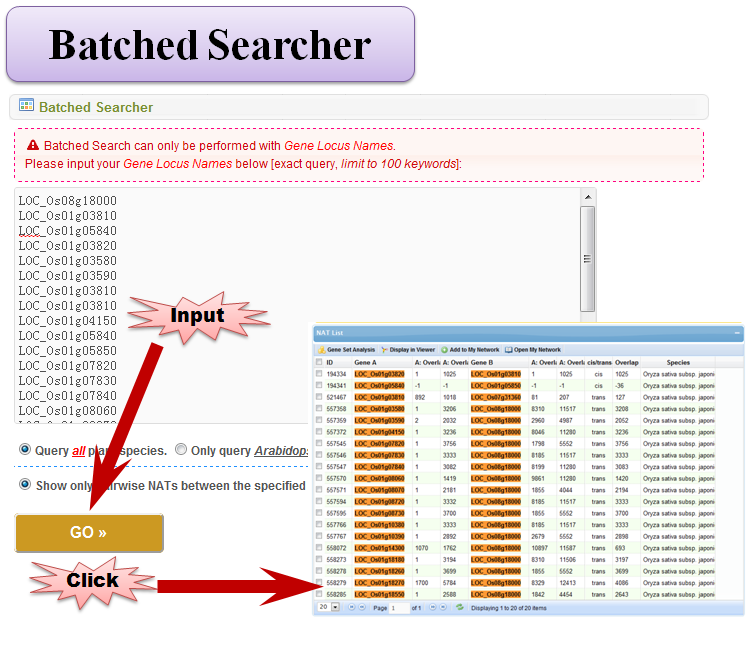
3.4 Batched Searcher 
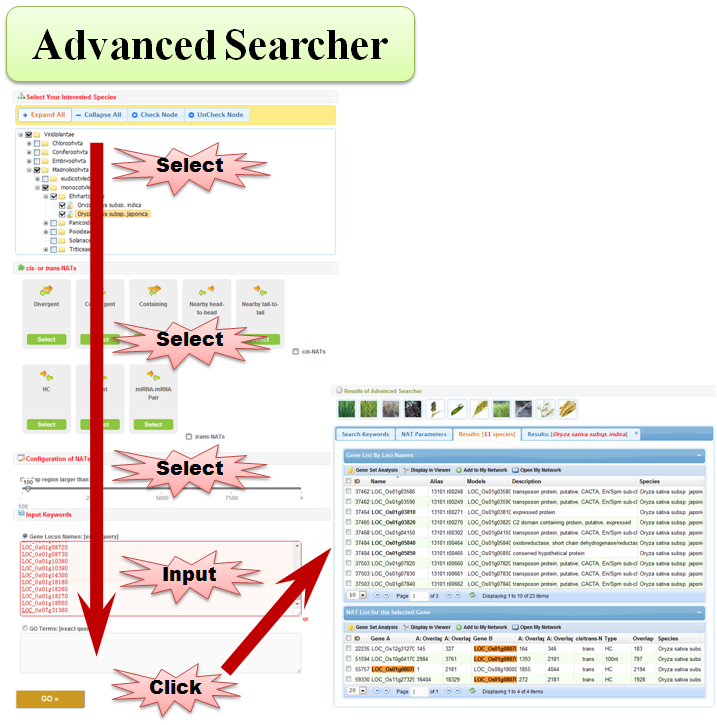
3.4 Advanced Searcher 
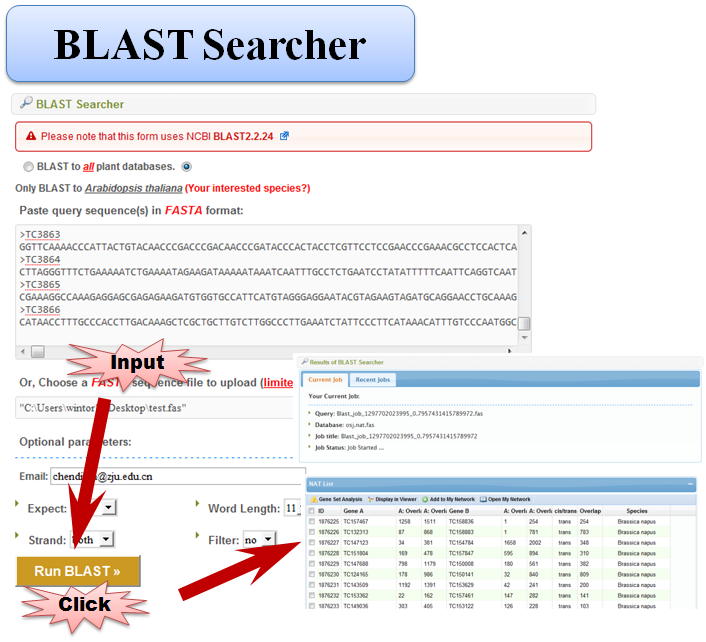
3.5 BLAST Searcher 
4 Viewer 
4.1 Gene Viewer 

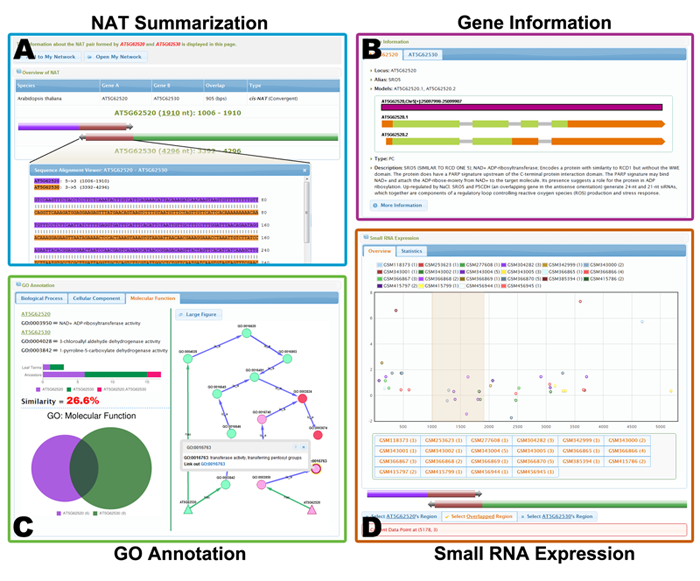
4.2 NAT Viewer 
The NAT page largely comprises four main parts, i.e., "NAT Summarization", "Gene Information", "GO Annotation" and "Small RNA Expression".
4.3 Network Viewer 
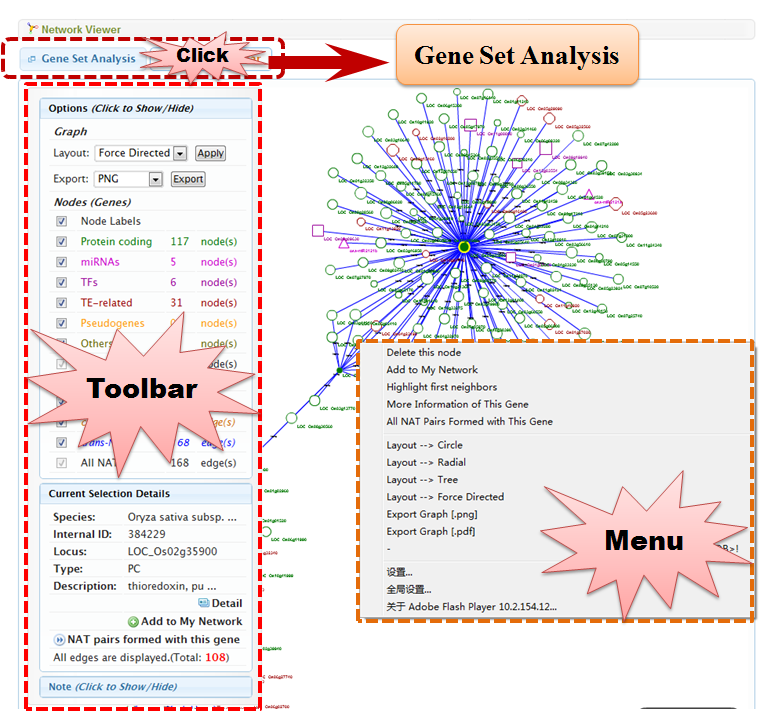
Network Viewer supports
5 Association Functions 
5.1 My Network 
In "My Network", genes or NAT pairs of interest may be stored temporarily on the server side during the session period and retrieved later. This feature will greatly facilitate users' digging of specific biological network formed by related NAT pairs involved in regulation of the same process. In many pages of the website, there is a button to add selected genes or NAT pairs to "My Network".
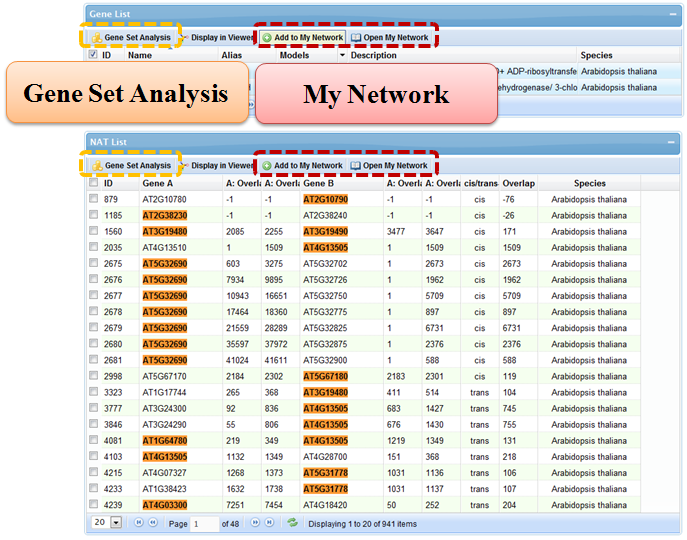
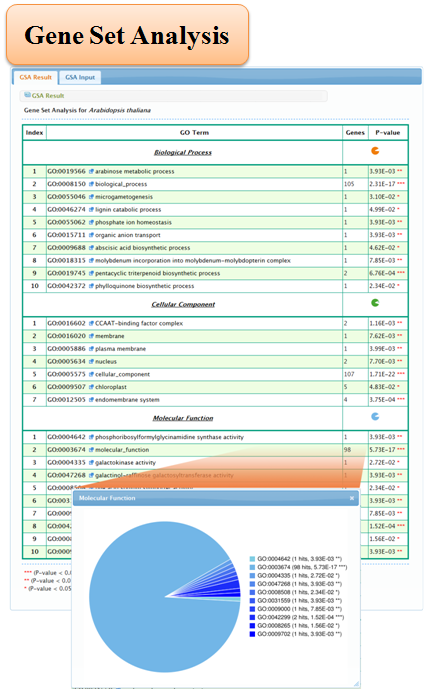
5.2 Gene Set Analysis 
6 Appendix 
6.1 Useful Hits 
It's recommended that you use Chrome, Firefox, Safari or Opera to access PlantNATsDB, although IE (Internet Explorer) or Netscape still work well.
6.2 Useful Links 
See the Useful Links tab of this page.
6.3 Methods 
See the Methods tab of this page.
6.4 References 
See the References tab of this page.
Methods:
Table of Contents
1. Prediction of NAT Pairs 
Prediction of NAT pairs was performed as previously described [17-18,22]. Specifically, the following criteria were used to identify cis-NATs and trans-NATs, respectively.
For cis-NATs, they can be grouped into five categories, namely, (i) Divergent (head to head or 5' to 5' overlap); (ii) Convergent (tail to tail or 3' to 3' overlap); (iii) Containing (full overlap); (iv) Nearby head-to-head (5' close to 5') and (v) Nearby tail-to-tail (3' close to 3') according to their relative orientation and degree of overlap (Figure 1A) [28]. If a pair of transcripts was located on opposite strands at adjacent genomic loci, and had at least a 1 nucleotide (nt) overlapping region or their distance on the chromosome was on longer than 100 nts, then they were considered as a cis-NAT pair. In total, 26 plant species were subjected to cis-NAT prediction.
For trans-NATs, BLASTN (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/, Release 2.2.20) [29] was used to search for transcript pairs with high sequence complementarity to each other and the following criteria should be satisfied for each transcript pair: (i) If the complementary region identified by BLAST covered more than half the length of either transcript, this transcript pair was designated to be a “high-coverage” (HC) trans-NAT pair; (ii) If the two transcripts had a continuous complementary region longer than 100 nts, they were classified as a “100 nt” pair. Functional trans-NATs should form RNA-RNA duplexes in vivo. We therefore used DINAMelt [30] to verify whether the transcript pairs could melt into RNA-RNA duplexes in the complementary regions in silico. All the trans-NAT pairs based on BLAST search were further used to DINAMelt hybridization validation. The trans-NAT pair was retained if it satisfied: (i) the paired region indentified by DINAMelt should be coincident with the BLAST-based search and (ii) any bubble in the paired region predicted by DINAMelt should be no longer than 10% of the region. For the BLAST-based trans-NAT pairs that contain transcripts longer than 10 Kb, they were not applied to DINAMelt validation due to the heavy computational work. Instead, it was considered as verified trans-NAT, if the paired region identified by BLAST was longer than 10% of its longer transcript.
All the NAT pairs predicted in this study were summarized in the Statistics page → Statistics of NATs tab.
2. Small RNA Analysis 
Small RNA (sRNA) sequences containing incomplete information (such as containing “N”) and with length less than 18 or more than 28 were removed for further analysis. For each data set, the filtered sRNA sequences were mapped to all the gene models of the related plant species. All mapping steps were performed using the Bowtie algorithm [33] allowing no mismatches. Besides, for comparison, the normalized abundance of sRNAs from each data set was calculated as RPMs (reads per million), which divided the read number of each sRNA by the total reads from this data set, and multiplied by 106.
For each NAT, an enrichment score was calculated to evaluate whether sRNAs were enriched in the overlapping region [17-18]. The enrichment score, E, was calculated using the following formula:
in which So = the total normalized abundance of the sRNAs generated from the overlapping region, Lo = the total length of the paired region of the two transcripts of the NATs, Sa = the total normalized abundance of the sRNAs generated from these two transcripts, and La = the total length of the two transcripts. Furthermore, a Pearson's chi-square test (χ2 test) was performed to test whether this enrichment was significant.
3. Gene Set Analysis 
Statistical tests that have been used for "Gene Set Analysis" to identify enriched GO categories include the Fisher's exact test, the χ2 test, the T test, the binomial test and the hypergeometric test. Here we used the combination of the χ2 test and Fisher's exact test to evaluate the significance of enrichment for GO category.
| Input Gene Set | Others | Total | |
|---|---|---|---|
| Interested Term | a | b | a+b |
| Other Terms | c | d | c+d |
| Total | a+c | b+d | a+b+c+d |
[1]. Fisher's exact test directly calculates the P-value using the following formula:
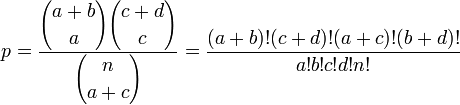
where 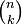 is the binomial coefficient and the symbol ! indicates the factorial operator.
is the binomial coefficient and the symbol ! indicates the factorial operator.
[2]. For χ2 test, The value of the test-statistic is

where:
- χ2 = Pearson's cumulative test statistic, which asymptotically approaches a χ2 distribution.
- Oi = an observed frequency (i.e., a, b, c and d in the above table);
- Ei = an expected frequency;
- n = the number of cells in the table (i.e., n = 4).
The chi-square statistic can then be used to calculate a P-value by comparing the value of the statistic to a χ2 distribution. The number of degrees of freedom df = 1.
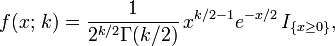
where:
- k = degrees of freedom df = 1
- Γ(k/2) denotes the Gamma function;.
- and,
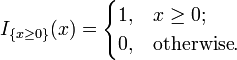
For small smaples (i.e., at least one of a, b, c and d is less than 5), PlantNATsDB uses the Fisher's exact test to calculate the P-value to evaluate the significance of enrichment for each category. With large samples, a χ2 test can be used in this situation.
References:
1. Ponting, C.P., Oliver, P.L. and Reik, W. (2009) Evolution and functions of long noncoding RNAs. Cell, 136, 629-641.
2. Brosnan, C.A. and Voinnet, O. (2009) The long and the short of noncoding RNAs. Curr Opin Cell Biol, 21, 416-425.
3. Ghildiyal, M. and Zamore, P.D. (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet, 10, 94-108.
4. Mercer, T.R., Dinger, M.E. and Mattick, J.S. (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet, 10, 155-159.
5. Lapidot, M. and Pilpel, Y. (2006) Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep, 7, 1216-1222.
6. Vanhee-Brossollet, C. and Vaquero, C. (1998) Do natural antisense transcripts make sense in eukaryotes? Gene, 211, 1-9.
7. Lavorgna, G., Dahary, D., Lehner, B., Sorek, R., Sanderson, C.M. and Casari, G. (2004) In search of antisense. Trends Biochem Sci, 29, 88-94.
8. Faghihi, M.A. and Wahlestedt, C. (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol, 10, 637-643.
9. Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R. and Zhu, J.K. (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell, 123, 1279-1291.
10. Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., Chiba, H., Kohara, Y., Kono, T., Nakano, T. et al. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature, 453, 539-543.
11. Tam, O.H., Aravin, A.A., Stein, P., Girard, A., Murchison, E.P., Cheloufi, S., Hodges, E., Anger, M., Sachidanandam, R., Schultz, R.M. et al. (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature, 453, 534-538.
12. Okamura, K., Balla, S., Martin, R., Liu, N. and Lai, E.C. (2008) Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol, 15, 998.
13. Czech, B., Malone, C.D., Zhou, R., Stark, A., Schlingeheyde, C., Dus, M., Perrimon, N., Kellis, M., Wohlschlegel, J.A., Sachidanandam, R. et al. (2008) An endogenous small interfering RNA pathway in Drosophila. Nature, 453, 798-802.
14. Ghildiyal, M., Seitz, H., Horwich, M.D., Li, C., Du, T., Lee, S., Xu, J., Kittler, E.L., Zapp, M.L., Weng, Z. et al. (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science, 320, 1077-1081.
15. Ron, M., Alandete Saez, M., Eshed Williams, L., Fletcher, J.C. and McCormick, S. (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev, 24, 1010-1021.
16. Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., Jr., Zhu, J.K., Staskawicz, B.J. and Jin, H. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A, 103, 18002-18007.
17. Zhou, X., Sunkar, R., Jin, H., Zhu, J.K. and Zhang, W. (2009) Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res, 19, 70-78.
18. Chen, D., Meng, Y., Ma, X., Mao, C., Bai, Y., Cao, J., Gu, H., Wu, P. and Chen, M. (2010) Small RNAs in angiosperms: sequence characteristics, distribution and generation. Bioinformatics, 26, 1391-1394.
19. Zhang, Y., Li, J., Kong, L., Gao, G., Liu, Q.R. and Wei, L. (2007) NATsDB: Natural Antisense Transcripts DataBase. Nucleic Acids Res, 35, D156-161.
20. Osato, N., Yamada, H., Satoh, K., Ooka, H., Yamamoto, M., Suzuki, K., Kawai, J., Carninci, P., Ohtomo, Y., Murakami, K. et al. (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol, 5, R5.
21. Wang, X.J., Gaasterland, T. and Chua, N.H. (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol, 6, R30.
22. Wang, H., Chua, N.H. and Wang, X.J. (2006) Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol, 7, R92.
23. Jin, H., Vacic, V., Girke, T., Lonardi, S. and Zhu, J.K. (2008) Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol, 9, 6.
24. Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 25, 25-29.
25. Lee, Y., Tsai, J., Sunkara, S., Karamycheva, S., Pertea, G., Sultana, R., Antonescu, V., Chan, A., Cheung, F. and Quackenbush, J. (2005) The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res, 33, D71-74.
26. Griffiths-Jones, S., Saini, H.K., van Dongen, S. and Enright, A.J. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res, 36, D154-158.
27. Edgar, R., Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res, 30, 207-210.
28. Osato, N., Suzuki, Y., Ikeo, K. and Gojobori, T. (2007) Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics, 176, 1299-1306.
29. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol, 215, 403-410.
30. Markham, N.R. and Zuker, M. (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res, 33, W577-581.
31. Allen, E., Xie, Z., Gustafson, A.M. and Carrington, J.C. (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 121, 207-221.
32. Pearson, W.R. and Lipman, D.J. (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A, 85, 2444-2448.
33. Langmead, B., Trapnell, C., Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10, R25.
34. Lopes, C.T., Franz, M., Kazi, F., Donaldson, S.L., Morris, Q. and Bader, G.D. (2010) Cytoscape Web: an interactive web-based network browser. Bioinformatics, 26, 2347-2348.
35. German, M.A., Pillay, M., Jeong, D.H., Hetawal, A., Luo, S., Janardhanan, P., Kannan, V., Rymarquis, L.A., Nobuta, K., German, R. et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol, 26, 941-946.
36. Addo-Quaye, C., Eshoo, T.W., Bartel, D.P. and Axtell, M.J. (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol, 18, 758-762.
Contact:
If you have any question/suggestion about PlantNATsDB, please feel free to contact us:
- Dijun Chen: chendijun@zju.edu.cn
- Chunhui Yuan: ch_yuan@zju.edu.cn
- Lingling Chen: llchen@mail.hzau.edu.cn
- Ming Chen: mchen@zju.edu.cn
Data Sources:
| ID | Scientific name | Common Name | Type a | Release Version | Release Date | Project Home | Download |
|---|---|---|---|---|---|---|---|
| aly | Arabidopsis lyrata | Lyrate rockcress | Chromosome | JGI Araly1 | Jul. 22, 2008 | Link | Link |
| ath | Arabidopsis thaliana | Arabidopsis | Chromosome | TAIR9 | Jun. 19, 2009 | Link | Link |
| bdi | Brachypodium distachyon | Purple false brome | Chromosome | JGI v1.0 8x | May 01 2009 | Link | Link Link |
| cpa | Carica papaya | Papaya | Scaffold | ASGPB v0.4 Dec 2 | Nov. 19, 2008 | Link | Link |
| cre | Chlamydomonas reinhardtii | - | Scaffold | JGI Chlre4 | Jan. 08, 2010 | Link | Link |
| csa | Cucumis sativus | Cucumber | Scaffold | JGI Cucsa_v1 | Jan. 08, 2010 | Link | Link |
| fve | Fragaria vesca | Strawberry | Scaffold | Version 2 | November 16, 2009 | Link | Link |
| gma | Glycine max | Soybean | Chromosome | JGI Glyma1 | Feb. 24, 2010 | Link | Link |
| lja | Lotus japonicus | Lotus | Chromosome | Lj 1.0 | May 18, 2009 | Link | Link |
| mac | Musa acuminata | dwarf banana | Chromosome | Version 1 | Aug. 9,2012 | Link | Link |
| mes | Manihot esculenta | Cassava | Scaffold | JGI Cassava1 | Nov. 09, 2009 | Link | Link |
| mgu | Mimulus guttatus | Spotted monkey flower | Scaffold | JGI Release v1.0 | Jan. 20, 2010 | Link | Link |
| mpu | Micromonas pusilla CCMP1545 | - | Scaffold | JGI Release v2.0 | Apr. 3, 2009 | Link | Link |
| msp | Micromonas sp. RCC299 | - | Scaffold | JGI Release v3.0 | Apr. 4, 2009 | Link | Link |
| mtr | Medicago truncatula | Medicago | Chromosome | Mt 3.0 | Oct. 12, 2009 | Link | Link |
| olu | Ostreococcus lucimarinus CCE9901 | - | Chromosome | JGI Release v2.0 | Oct. 27, 2007 | Link | Link |
| osi | Oryza sativa subsp. indica | Rice (indica) | Chromosome | BGI Release | Oct. 28, 2008 | Link | Link |
| osj | Oryza sativa subsp. japonica | Rice (japonica) | Chromosome | TIGR Rice Release 6.1 | Jun. 3, 2009 | Link | Link |
| ota | Ostreococcus tauri | - | Chromosome | JGI Release v2.0 | Apr. 11, 2008 | Link | Link |
| ppa | Physcomitrella patens | Moss | Scaffold | JGI Phypa1.1 | Mar. 22, 2007 | Link | Link |
| ppe | Prunus persica | Peach | Scaffold | JGI v1.0 | May 26, 2009 | Link | Link |
| ptr | Populus trichocarpa | Poplar | Chromosome | JGI Ptr v2.0 | Mar. 16, 2010 | Link | Link |
| rco | Ricinus communis | Castor bean | Scaffold | TIGR/JCVI Release v0.1 | May 22, 2008 | Link | Link |
| sbi | Sorghum bicolor | Sorghum | Chromosome | JGI Sbi1 | Mar. 25, 2008 | Link | Link |
| smo | Selaginella moellendorffii | Spikemosses | Scaffold | JGI Selmo1 | Oct. 19, 2007 | Link | Link |
| vca | Volvox carteri | Volvox | Scaffold | JGI Release v2.0 | Sep. 13, 2007 | Link | Link |
| vvi | Vitis vinifera | Grape | Chromosome | Genoscope12x | Mar. 19, 2010 | Link | Link |
| zma | Zea mays | Maize | Chromosome | Release 4a.53 | Mar. 20, 2009 | Link | Link |
| ace | Allium cepa | Onion | TC | OnGI Release 2.0 | Jul. 17, 2008 | Link | Link |
| aco | Aquilegia coerulea | Aquilegia | TC | AqGI Release 2.1 | Jun. 6, 2008 | Link | Link |
| bna | Brassica napus | Rapeseed | TC | BnGI Release 4.0 | Mar. 16, 2010 | Link | Link |
| bvu | Beta vulgaris | Beet | TC | BvGI Release 3.0 | Jun. 16, 2010 | Link | Link |
| can | Capsicum annuum | Pepper | TC | PepGI Release 4.0 | May 21, 2009 | Link | Link |
| cca | Coffea canephora | Coffee | TC | CocaGI Release 2.0 | Apr. 14, 2010 | Link | Link |
| ccl | Citrus clementina | Clementine | TC | CiclGI Release 2.0 | May 22, 2009 | Link | Link |
| ees | Euphorbia esula | Leafy spurge | TC | EuesGI Release 1.0 | Jun. 30, 2008 | Link | Link |
| esi | Ectocarpus siliculosus | Brown algae | TC | BaGI Release 1.0 | May 27, 2010 | Link | Link |
| far | Festuca arundinacea | Tall fescue | TC | FaGI Release 3.0 | Apr. 7, 2010 | Link | Link |
| fpr | Festuca pratensis | Meadow ryegrass | TC | MrGI Release 1.0 | May 28, 2010 | Link | Link |
| ghi | Gossypium hirsutum | Upland cotton | TC | CGI Release 10.1 | Mar. 4, 2010 | Link | Link |
| gra | Gossypium raimondii | Cotton | TC | GoraGI Release 1.0 | Jul. 2, 2008 | Link | Link |
| han | Helianthus annuus | Sunflower | TC | HaGI Release 6.0 | May 24, 2009 | Link | Link |
| hvu | Hordeum vulgare | Barley | TC | HvGI Release 11.0 | Apr. 14, 2010 | Link | Link |
| ini | Ipomoea nil | Morning glory | TC | IpniG Release 1.0 | Jun. 30, 2008 | Link | Link |
| lsa | Lactuca sativa | Garden lettuce | TC | LsGI Release 3.0 | Jul. 2, 2008 | Link | Link |
| lse | Lactuca serriola | Prickly lettuce | TC | LaseGI Release 1.0 | Jun. 28, 2008 | Link | Link |
| mcr | Mesembryanthemum crystallinum | Ice plant | TC | McGI Release 5.0 | Jun. 19, 2008 | Link | Link |
| mdo | Malus x domestica | Apple | TC | MdGI Release 3.0 | Apr. 8, 2010 | Link | Link |
| nbe | Nicotiana benthamiana | - | TC | NbGI Release 4.0 | Apr. 9, 2010 | Link | Link |
| nta | Nicotiana tabacum | Tobacco | TC | NtGI Release 6.0 | Apr. 10, 2010 | Link | Link |
| pco | Phaseolus coccineus | Scarlet bean | TC | PcGI Release 1.0 | May 27, 2009 | Link | Link |
| phy | Petunia hybrida | Petunia | TC | PhGI Release 1.0 | Apr. 14, 2010 | Link | Link |
| pta | Pinus taeda | Loblolly pine | TC | PGI Release 8.0 | Apr. 15, 2010 | Link | Link |
| pvi | Panicum virgatum | Switchgrass | TC | PaviGI Release 1.0 | May 28, 2009 | Link | Link |
| pvu | Phaseolus vulgaris | Kidney bean | TC | PhvGI Release 1.0 | Apr. 15, 2010 | Link | Link |
| qro | Quercus robur | Oak | TC | OGI Release 1.0 | Jul. 1, 2010 | Link | Link |
| rsa | Raphanus sativus | Radish | TC | RsGI Release 1.0 | May 27, 2010 | Link | Link |
| sce | Secale cereale | Rye | TC | RyeGI Release 4.0 | Jul. 3, 2008 | Link | Link |
| she | Striga hermonthica | Purple witch weed | TC | ShGI Release 1.0 | May 27, 2010 | Link | Link |
| sly | Solanum lycopersicum | Tomato | TC | LeGI Release 13.0 | Apr. 13, 2010 | Link | Link |
| sme | Solanum melongena | Eggplant | TC | SomeGI Release 1.0 | May 28, 2010 | Link | Link |
| sof | Saccharum officinarum | Sugarcane | TC | SoGI Release 1.0 | Apr. 9, 2010 | Link | Link |
| stu | Solanum tuberosum | Potato | TC | StGI Release 13.0 | Apr. 16, 2010 | Link | Link |
| tae | Triticum aestivum | Wheat | TC | TaGI Release 12.0 | Apr. 18, 2010 | Link | Link |
| tca | Theobroma cacao | Cocoa | TC | TcaGI Release 3.0 | May 21, 2009 | Link | Link |
| ter | Triphysaria eriantha | Triphysaria | TC | TriphGI Release 1.0 | Aug. 15, 2008 | Link | Link |
| tve | Triphysaria versicolor | - | TC | TverGI Release 2.0 | Jun. 28, 2008 | Link | Link |
| vun | Vigna unguiculata | Cowpea | TC | VuGI Release 1.0 | May 27, 2010 | Link | Link |
| pab | Picea abies | Spruce | TC | Sgi Release 4.0 | Mar. 5, 2010 | Link | Link |
| csi | Citrus sinensis | Orange | TC | CsGI Release 1.0 | Jun. 25, 2008 | Link | Link |
aTCs: "Tentative Consensus" sequences, are assemblies of ESTs.
Plant Information Resource
- TAIR: The Arabidopsis Information Resource
- ASRP: Arabidopsis small RNA project
- AtGenExpress: a multinational effort to uncover the transcriptome of A. thaliana.
- Arabidopsis epigenome maps
- CATdb: A Complete Arabidopsis Transcriptome database
- PlantTFDB: Plant Transcription Factor Database
- PlnTFDB: The Plant Transcription Factor Database
- The TIGR Rice Genome Annotation Project
- RISE: Beijing Genome Institute Rice Information System
- RAP-DB: Rice Annotation Project Database
- the Rice Array Database
- Plant MPSS (Massively Parallel Signature Sequencing) Database: datasets for Arabidopsis, rice, maize, Brachypodium, Legume, and grape, at U Delaware
- Rice MPSS data at U. Delaware
- Arabidopsis MPSS data at U Delaware
- Grape MPSS data at U Delaware
- Maize genome sequence
- MaizeDB
- Papaya Genome Project at University of Hawaii
- Castor Bean Genome Database at TIGR
- Lotus japonicus genome project at Kazusa
- Soybean genome map at Southern Illinois University
- Phytozome: Comparative Genomics of Plants.
- DFCI - Plant Gene Indices: The Plant Gene Index Project
- Gramene: a Resource for Comparative Grass Genomics
- PMRD: plant microRNA database
- miRBase: the microRNA database
- Comparative Sequencing of plant small RNAs
- CSRDB: a small RNA integrated database and browser resource for cereals
- GEO: Gene Expression Omnibus
- PlantGDB: Plant Genome Database
- NCBI Plant Genomes Central: Links to NCBI resources for plant genomics.
- Soybean genome sequence at Phytozome
- Medicago truncatula: a model for legume research
- Legume Information System
- Soybase and the soybean breeders toolbox
- Lotus japonicus genome project at Kazusa
- BeanGenes: a Phaseolus/Vigra database, including links to other useful sites.
- SoyMap: An integrated map of soybean for resolution and dissection of multiple genome duplication events.
- Soybean genome map at Southern Illinois University
- the MIPS plant genomics group: focus on the analysis of the Arabidopsis, maize, Medicago truncatula, Lotus, rice, tomato, sorghum, barley and other plant genomes.
- PLAZA: Green plant comparative genomics
- AGI: Arizona Genome Institute
- SIGnAL: Salk Institute Genomic Analysis Laboratory
- CUGI: Clemson University Genomics Institute
- ChromatinDB
- Carbohydrate active enzymes
- Cell Wall Genomics
- Primer3 primer design tool
- Gene Ontology
- Plant Ontology
- GRIN: Germplasm Resources Information Network
- The Virtual Plant at NYU.
- Agricola, a Bibliographic database of citations to the agricultural literature
- Phytome supports phylogenetic and functional analyses of predicted protein sequences across plants.
- OrthologID provide phylogenetic analysis of Arabidopsis, rice, Populus, and Chlamydomonas genes
- PlantTribes provides OrthoMCL clustering of plant proteins, developed as part of the Floral Genome Project.
Author: Dijun Chen © 2010-2012 Ming Chen's Lab, Zhejiang University