Document
We screen all of the literature in the PubMed database with following keywords: circular RNA, circRNA or RNA circularization. The relevant hits were downloaded and further inspected manually. We extracted information on functional circRNAs, their related diseases and biological regulations.
The GO annotations were downloaded from the Gene Ontology Consortium. Besides, the RBPs matching to human functional circRNAs were from CircInteractome. Mature miRNA sequences were acquired from miRBase, while animal and plant miRNA-circRNA interactions were predicted by miRanda and TargetFinder, respectively.
Currently, a user-friendly search interface is provided at the Search page. Users can retrieve the database by circRNA name, circRNA location, gene symbol or keywords (such as 'lung cancer' for human). Functional circRNAs from eight animal and six plant species are available in the current version of CircFunBase.
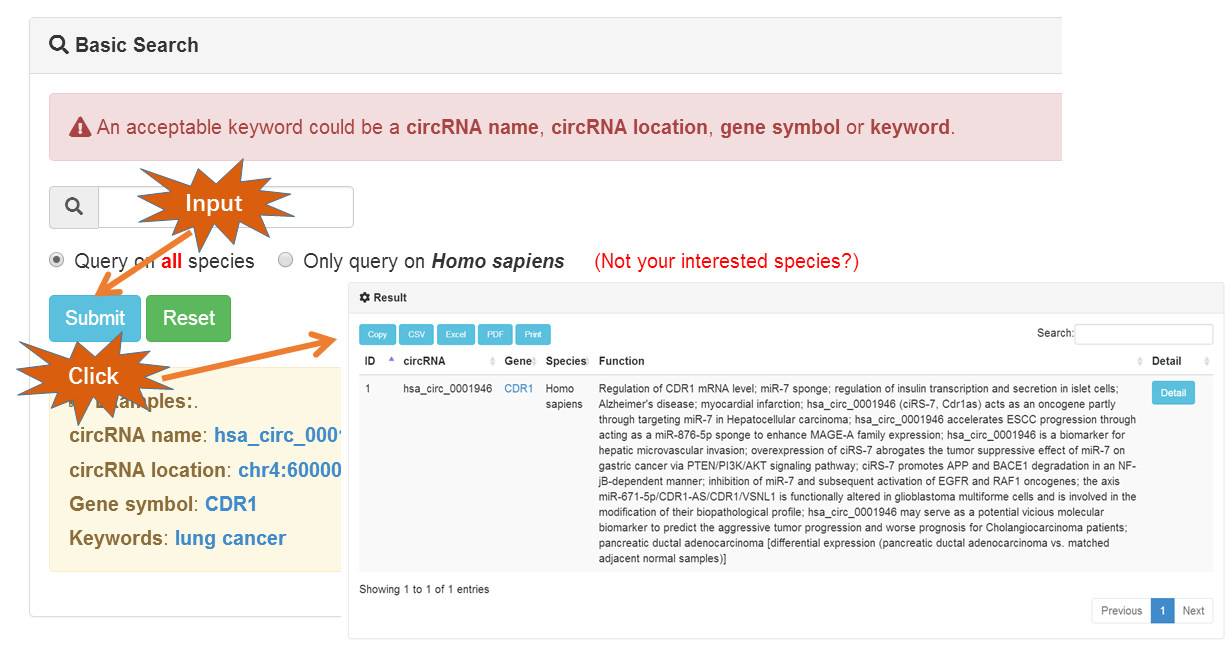
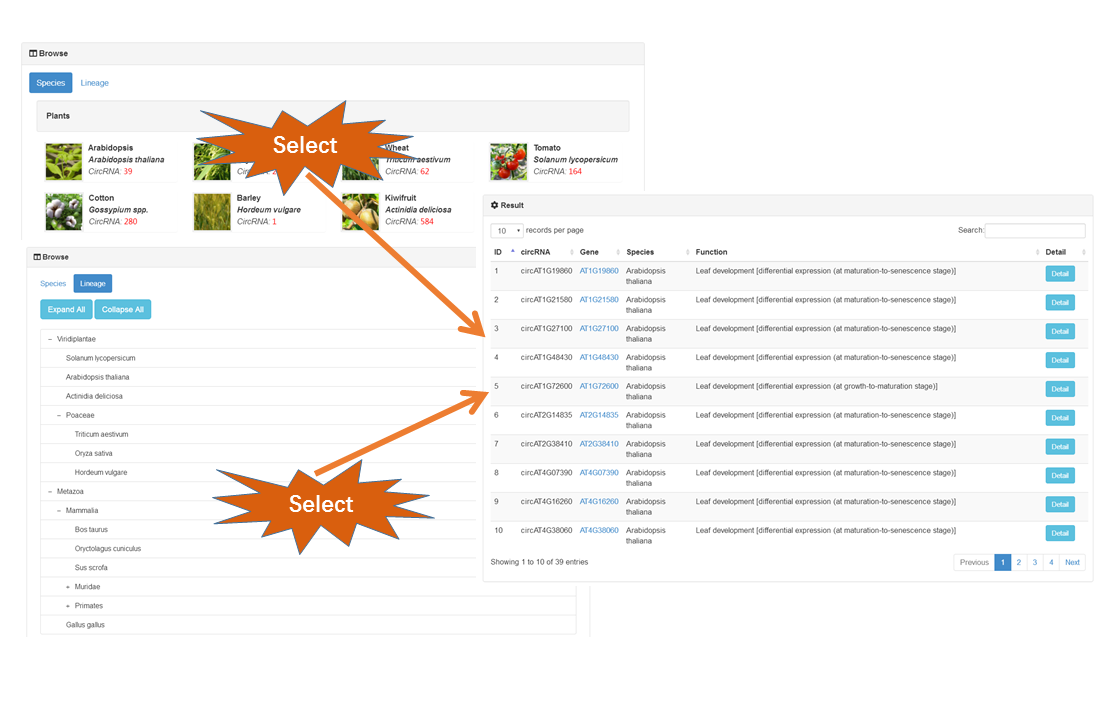
In "Browse" page, users can browse the circRNAs by species or by lineage to have the general info of interested organism. In addition, each column of the table can be sorted. If you want to download all the circRNAs infomation, please the Download part.
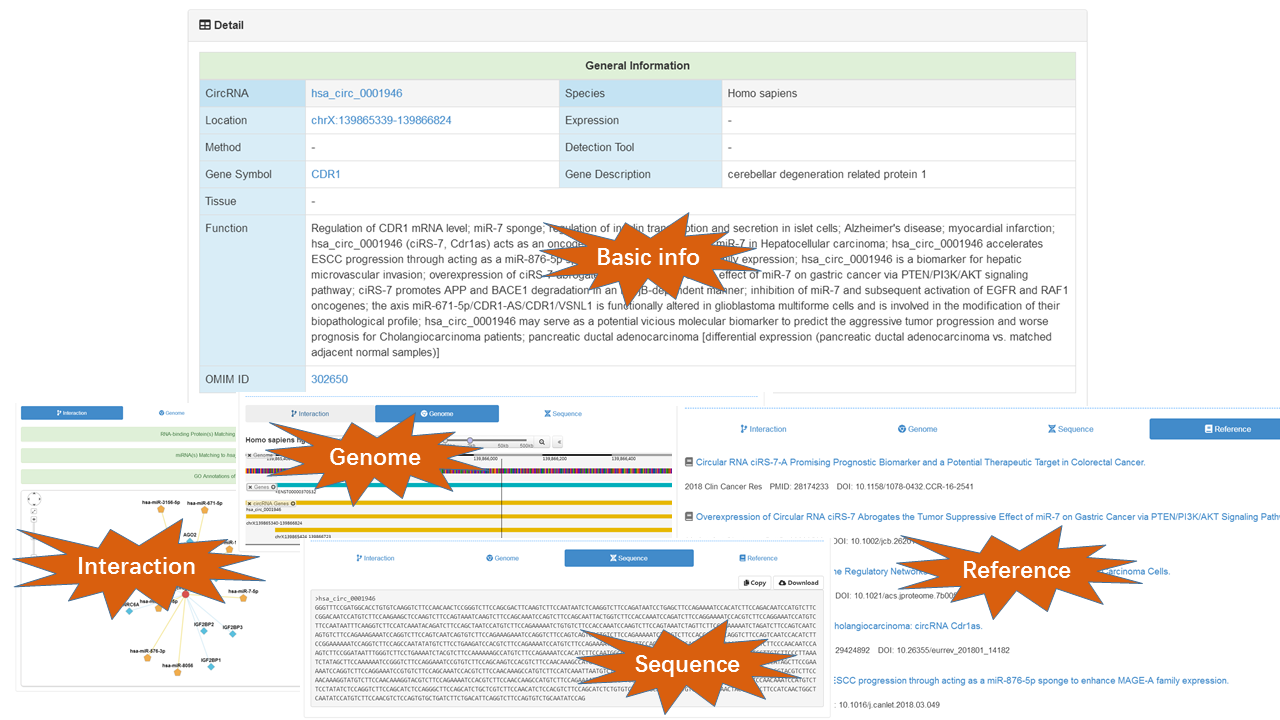
CircFunBase provides brief description of search results in the "Search Result" page or "Browse Result" page. To gain more detail information of a specific circRNA, users can click the "Details" button. Additional information such as PubMed ID, GO annotations and circRNA-associated miRNAs are displayed in the circRNA single-record page. We also provide many useful hyperlinks: 1) human and mouse circRNAs are linked to circBase and could be viewed in UCSC Genome Browser; 2) gene symbol is linked to the NCBI Gene (for Solanum lycopersicum, gene symbol is linked to Ensembl); 3) clicking the GO term links to the GO Consortium website; 4) miRNA name is linked to mirBase. Together, these information will help users to understand circRNA functions.
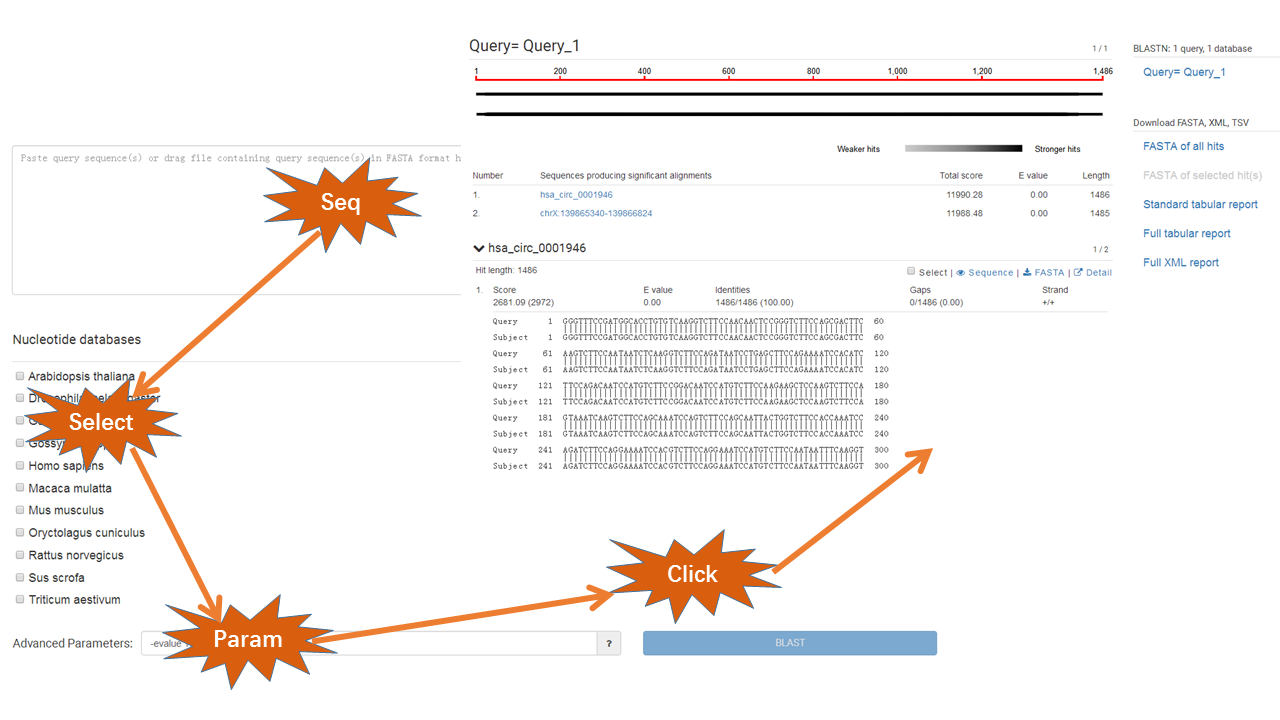
CircFunBase provides blast module to help interpret better unknown sequences by alignment against with knwon functional circRNAs sequences. And custom paramters and specific species databases can be defined. The result page shows detailed alignment infomation and can be redirected to the detailed information page.
Functional circRNA list of each species can be downloaded here.
Animals :
Plants :
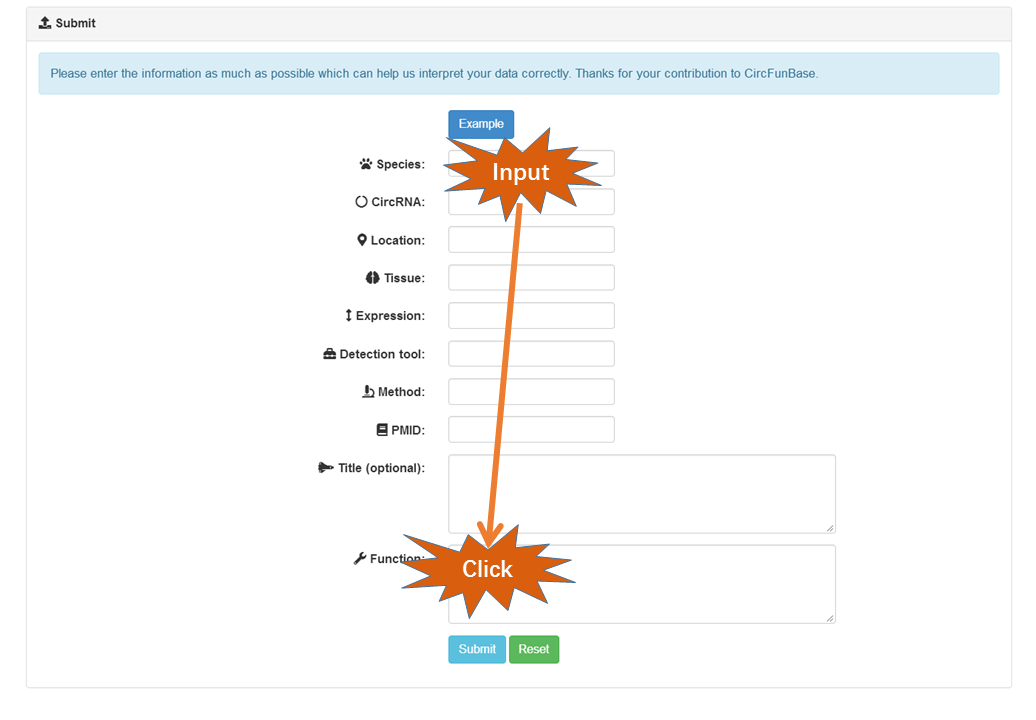
It is inevitable that the collections of CircFunBase may not cover all functional circRNAs. So we provide the submission interface to make sure that researchers can submit novel functional circRNAs that are not documented. In addition, multiple circRNA entries could be submitted in a file.
CircFunBase provides a series of APIs to return detailed information in JSON format, for example, circRNA information or miRNA interaction information. Researchers can refer the API documentations for usage.
If you have any question/suggestion about CircFunBase, please feel free to contact us.
Contact
Ming Chen, Professor, College of Life Sciences, Zhejiang University.
Address
Department of Bioinformatics, College of Life Sciences, Yuhangtang Road 866, Xihu District, Hangzhou, Zhejiang, P. R. China, 310058.
Developers