What's dscBLAST
dscBLAST: developmental single-cell Basic Local Alignment Search Tool
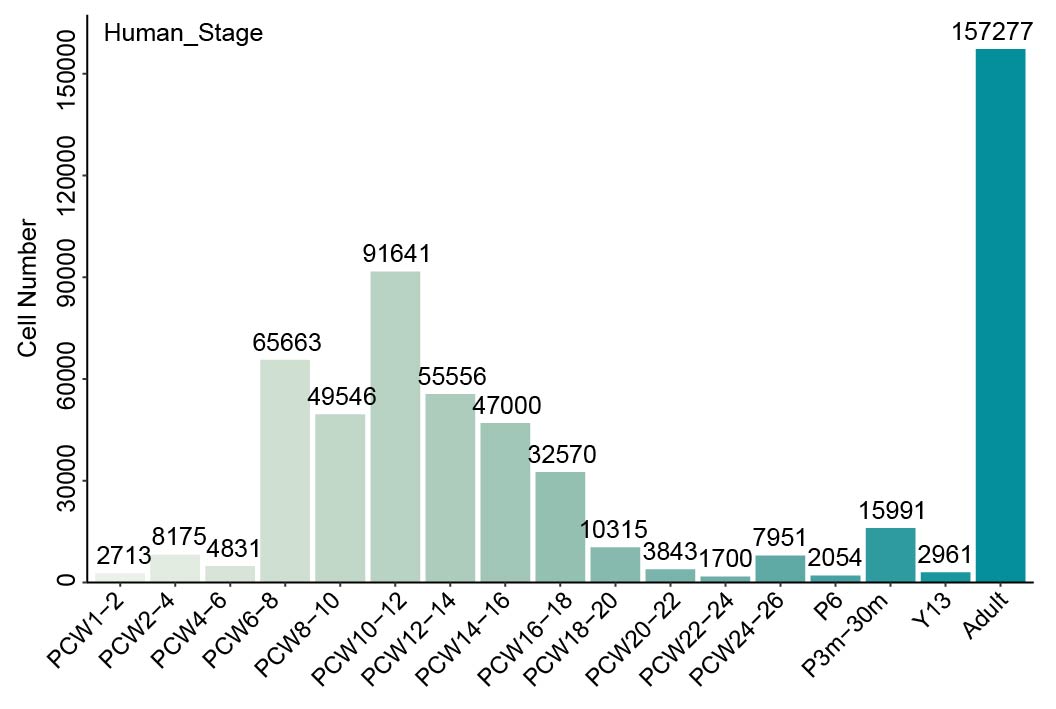
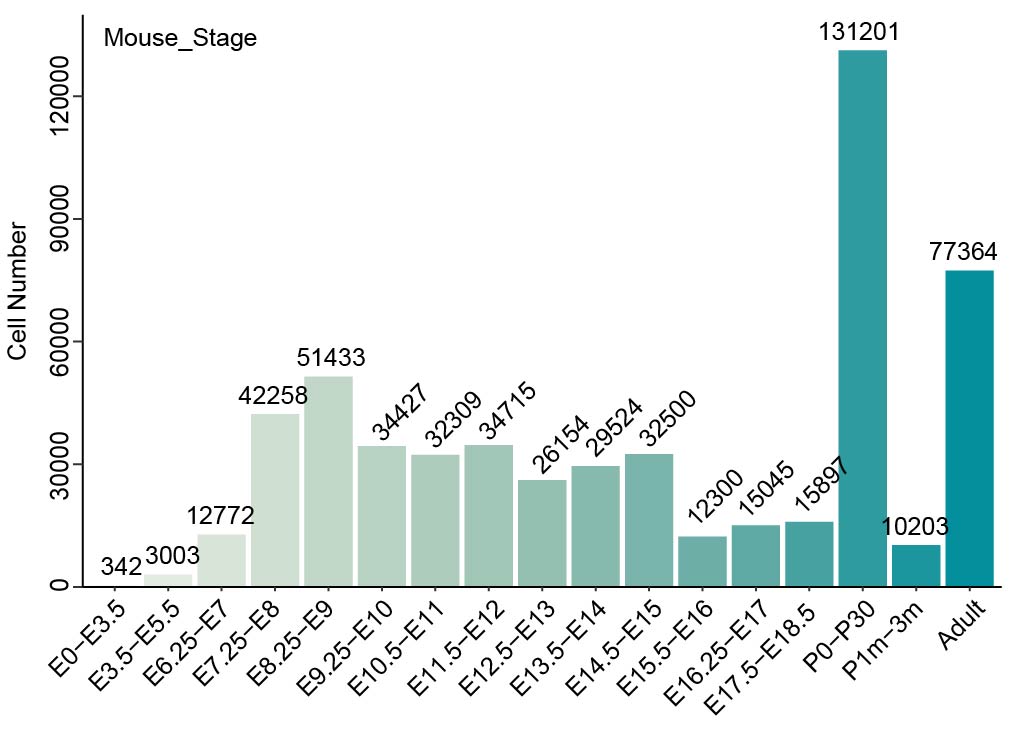
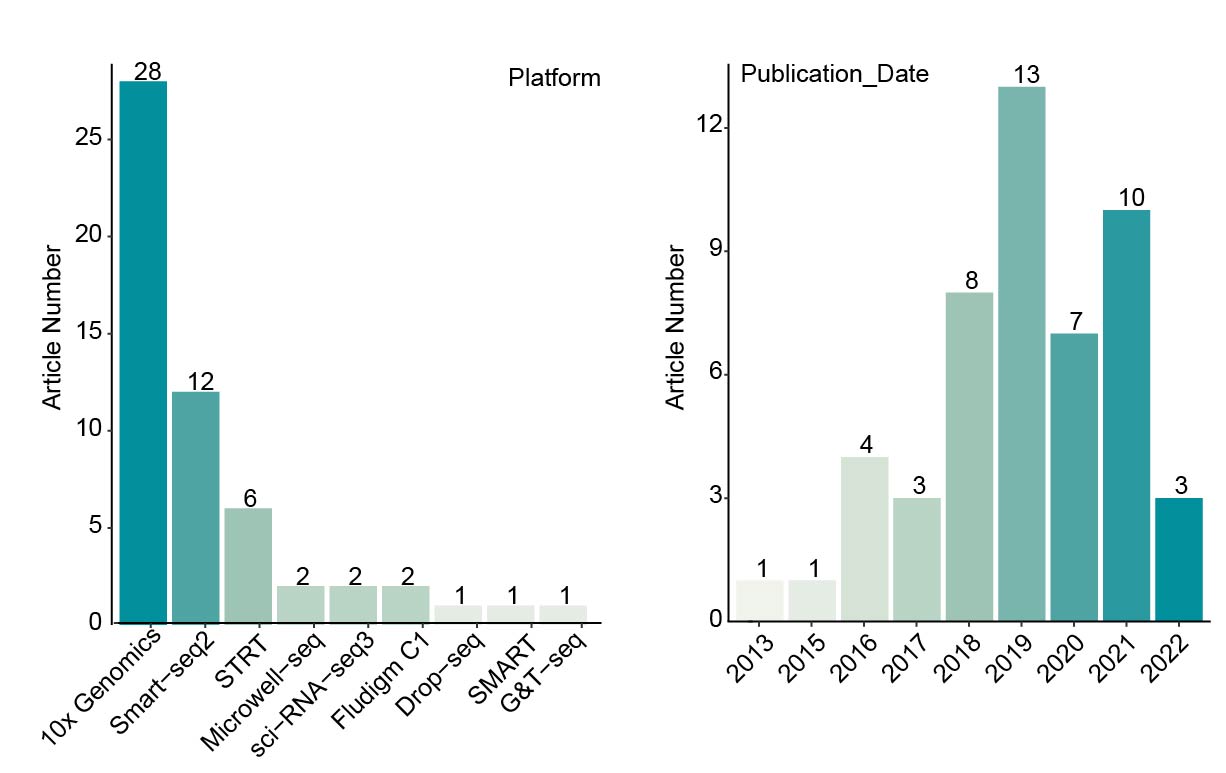
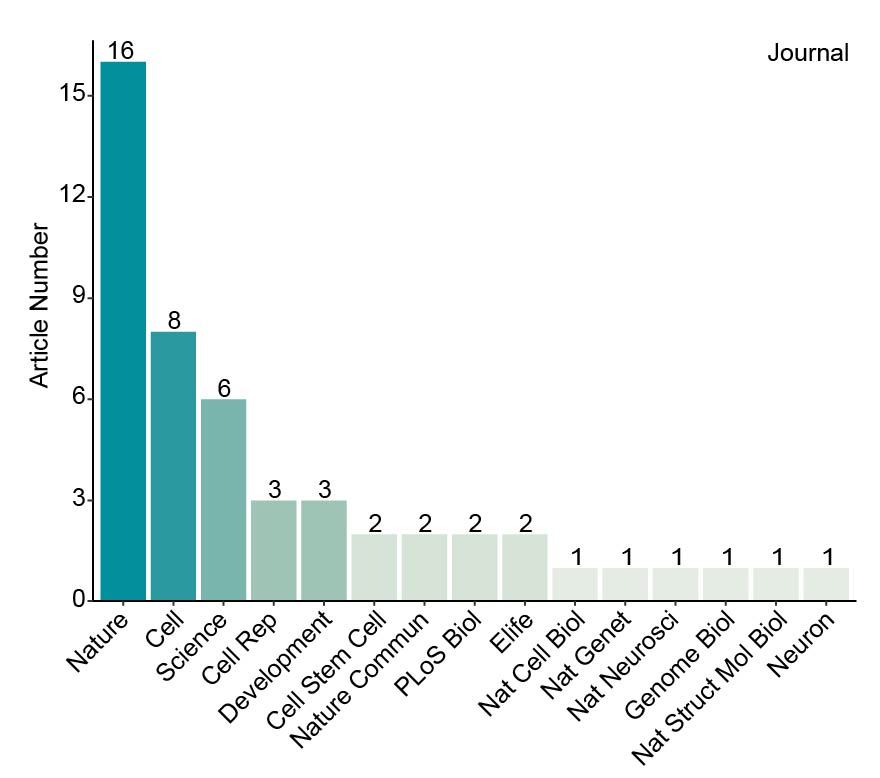
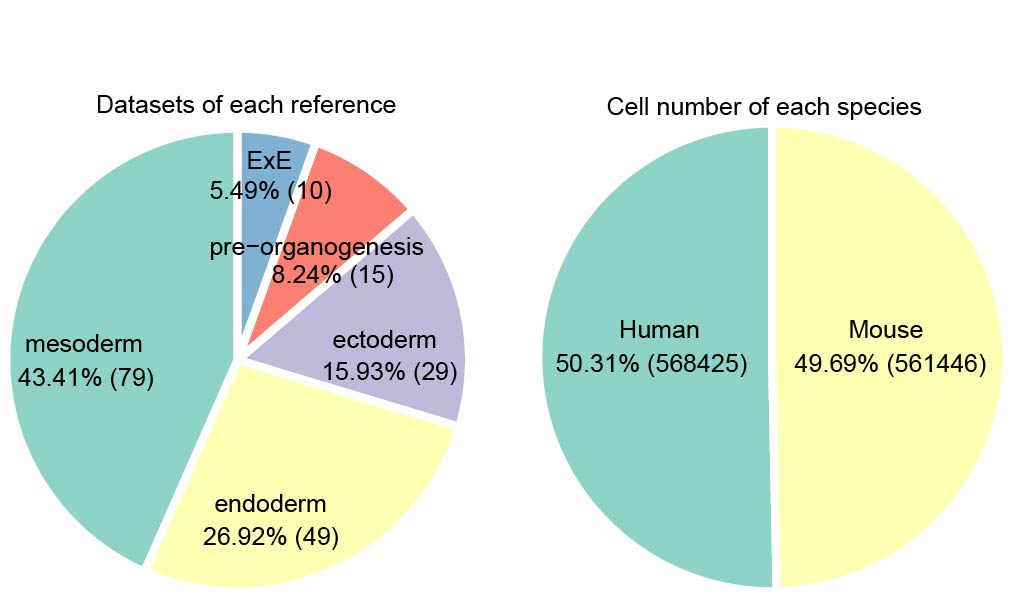
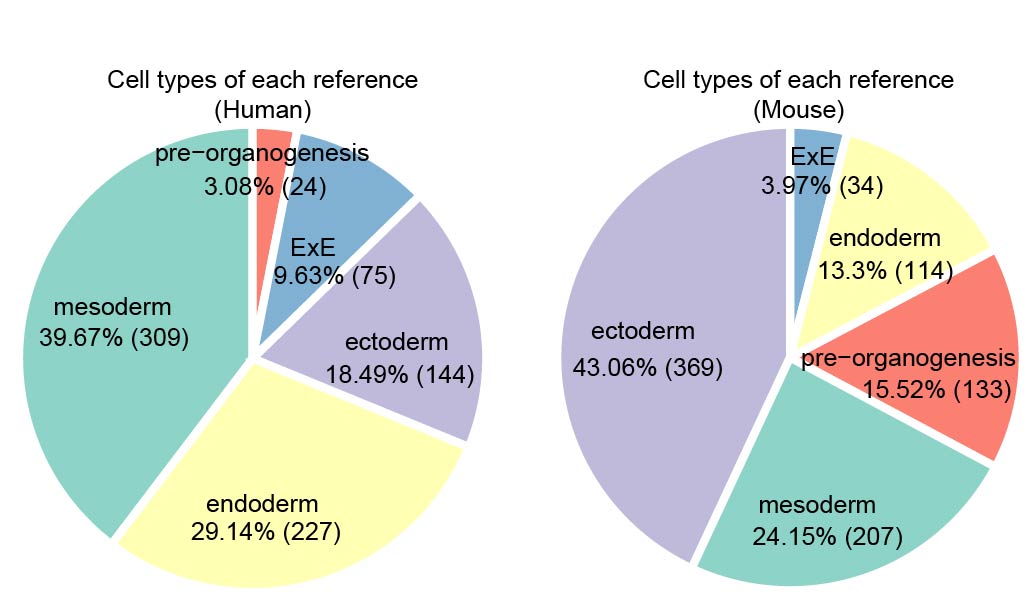
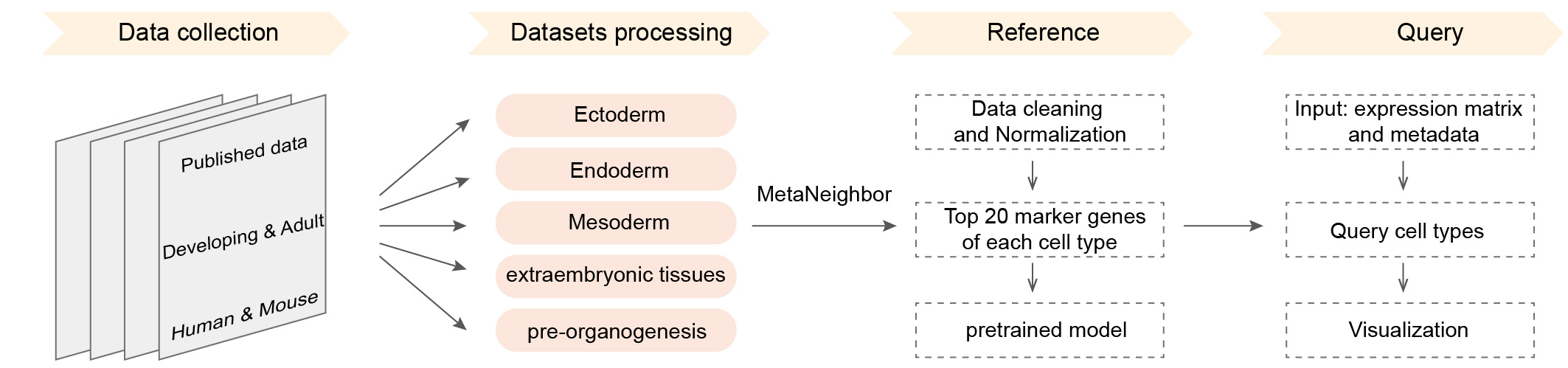
We collected more than 160 datasets (Organ_First author) from approximately 50 articles. These data were grouped into five references: Ectoderm, Endoderm, Mesoderm, pre-organogenesis and extraembryonic tissues. Then MetaNeighbor was used to calculate the similarity between query cells and reference cells. The combination of comprehensive references and MetaNeighbor enables the users to confirm the cell identity and speculate its possible developmental stages rapidly and accurately. The dscBLAST is particularly better at developmental data, and it also performs very well on adult data.
RUN dscBLAST
R package dscBLAST is supported to annotate cell types and speculate the developmental stages of cells. An exhaustive help document is available at our Github: https://github.com/Fuyt27/dscBLAST
Workflow
References Summary
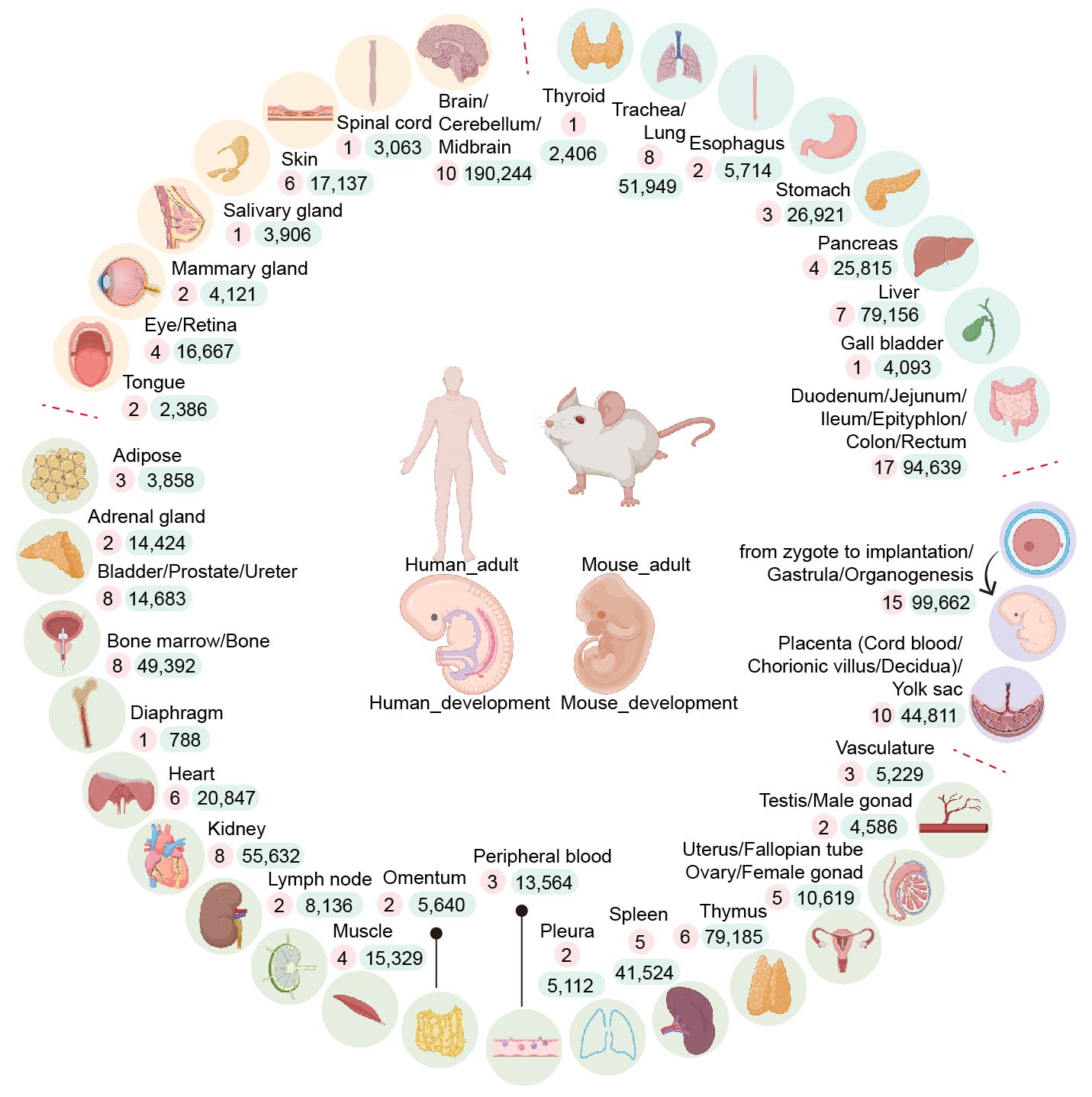
Summary of collected organs and their datasets.
Yellow background: Organs from ectoderm;
Green background: Organs from mesoderm;
Blue background: Organs from endoderm;
Purple background: Organs or structures from extraembryonic tissues or pre-organogenesis stages;
Numbers with red and round background: The number of datasets included in their corresponding organs;
Numbers with blue and rounded rectangle background: The cell number of each organ.
Created by BioRender.
Citing dscBLAST
Daiyuan Liu et al., Characterization of human pluripotent stem cell differentiation by single-cell dual-omics analyses. Stem Cell Reports. 2023.